Experimental animals
Experiments using animals were performed in accordance with the guidelines set forth by the National Institutes of Health and approved by the National Institute of Neurological Disorders and Stroke or the National Institute of Dental and Craniofacial Research Animal Care and Use Committees. Mouse lines Ai95(RCL-GCaMP6f)-D (no. 024105)30, Mrgprd–CreERT2 (no. 031286)29, Mrdprb4-tdT-2a–Cre (no. 021077)4, Trpv1–Cre (no. 017769)54, Sst–Cre (no. 013044)55 and Trpv1−/− (no. 003770)38 were purchased from The Jackson Laboratory. Tac1–tagRFP–2a–TVA mouse line was described previously56, and the Avil–Flp line57 was a gift from D. Ginty. We also generated a new Rosa–Cag–LSL–soma–jGCaMP8s line used in crosses with Sst–Cre, an Avil–LSL–2A–TeNT line and a Rosa–CAG–FSF–LSL–KORD line using CRISPR–Cas9-mediated recombination56. Male and female mice were used in all experiments and given ad libitum access to standard laboratory chow and water. The mice were housed in a controlled environment (23 °C and 50% humidity with a 12-h light–dark cycle); no statistical methods were used to predetermine sample size.
Injection of adeno-associated virus in mouse pups and CreERT2 induction
Left intracerebroventricular injection of 1 μl of AAV9-Cag–Cre virus (2 × 1012 to 2 × 1013 virions ml−1; catalogue no. CV17187-AAV9; Vigene) or AAV9-CMV–Cre virus (more than 1 × 1013 virions ml−1; catalogue no. 105537-AAV9; Addgene) in 1- to 3-day-old mouse pups containing Ai95 was used to achieve stochastic expression of GCaMP in neurons of the trigeminal ganglion as described previously42. To induce CreERT2 recombination, tamoxifen (Sigma-Aldrich) was dissolved in corn oil at a concentration of 20 mg ml−1 at 37 °C overnight. Mrgprd–CreERT2 mice were injected intraperitoneally at a dosage of 75 mg tamoxifen per kg of body weight using an insulin syringe at least a week before calcium imaging.
In vivo calcium imaging
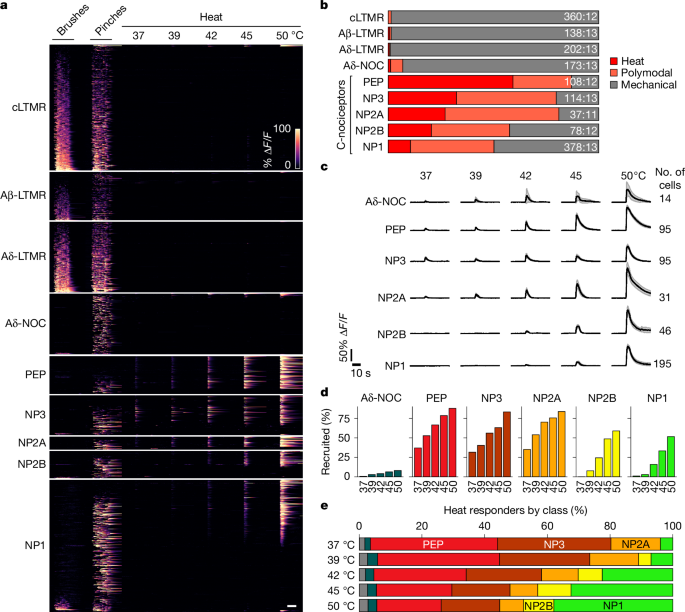
For fluorescent calcium imaging of the trigeminal neurons, adult (over 8 weeks old) animals were subjected to one of three experimental regimes. Experimental regime A assessed both mechanical and temperature responses (Figs. 1 and 2 and Extended Data Figs. 1–5). The animals were anaesthetized with isoflurane and surgically prepared for optical access to the trigeminal ganglion as described previously19. The hairy skin of the cheek was mechanically stimulated using a series of manually delivered brushes with a cotton-tipped applicator and pinches with surgical forceps as described previously3. For improved temperature transfer, the mouse cheek was treated for less than 300 s with a depilatory cream (Veet) using a cotton-tip applicator. An eye ointment was applied to prevent damage to the cornea and conjunctiva, fur was removed, the cheek was washed at least three times with saline and dried with Kimwipes and a custom-built 5 mm2 Peltier probe (TCS2; QST.Lab) was applied directly to the skin for thermal stimulation. Peltier application did not induce long-term activity of trigeminal neurons, and LTMRs did not respond to heating (Fig. 1), ruling out major confounding effects related to mechanical stimulation by the probe. The skin was held at 30 °C for baseline non-stimulated activity measurements; temperature stimulation at 37, 39, 42, 45 and 50 °C was for 4 s.
Experimental regime B assessed the mechanical sensitization after inflammation (Fig. 4 and Extended Data Fig. 7). Baseline mechanical stimulation was as in experimental regime A. Inflammation of the cheek was then induced using subdermal injection of 20–30 μl, 0.5 mM PGE2 (Sigma-Aldrich) to three sites. After 10 min of incubation, inflammation was confirmed and mechanical stimulation was repeated.
Experimental regime C assessed the thermal sensitization after inflammation (Figs. 3 and 5 and Extended Data Figs. 5 and 6). Mouse cheek depilation was carried out the day before functional imaging. Skin was subjected to a series of pinches and thermal stimulation as in experimental regime A. PGE2 or LY344864 (20–30 μl; 5 mg ml−1; MilliporeSigma) was injected as in experimental regime B, and stimulation was repeated. To evaluate consistency, responses to common stimuli were compared between experimental regimes A and C (Extended Data Fig. 10).
The functional activity of DRG neurons in the L5 and L6 ganglia was determined using fluorescent calcium imaging. PGE2 and LY344864 were injected as a single injection (volumes and concentrations as above) into the plantar hind paw. Mechanical stimulation was also applied to the plantar surface, which was held at 30 °C for the unstimulated recordings. For Mrgprb4-tdT-2a–Cre mice, the hairy skin of the leg was stimulated as NP2B neurons selectively innervated hairy skin4.
Calcium imaging was performed as described previously3,58 using a custom-built epifluorescence Cerna microscope (Thorlabs) and a pco.panda 4.2 bi CMOS camera; 40-s recording episodes were acquired at 5 Hz. For each experiment requiring post hoc ISH, either red fluorescent tagRFP images were collected or the trigeminal ganglion was briefly superfused with 500 μl of 1 M KCl to activate and visualize all GCaMP-expressing neurons after the experiment to provide alignment guide-posts. In vivo images were aligned and processed as described previously3.
Spatial activity maps and analysis of fluorescence dynamics
Spatial activity maps and regions of interest (ROI) were generated as described previously3. In brief, activity induced by repetitive mechanical stimulation was visualized as standard deviation over time for each pixel. Heat-induced activity was visualized by subtracting the mean fluorescence before stimulation from the mean fluorescence during stimulation. ROI were manually extracted using the ‘Cell Magic Wand’ plugin in ImageJ. Overlapping cell ROI that were contaminated by each other’s responses were excluded from the analysis while blind to transcriptomic information. Relative change in GCaMP fluorescence was calculated as ΔF/F (%) for each cell, and potential contaminant signal from the underlying out-of-focus tissue and neighbouring cells was removed by subtracting the fluorescence of a doughnut-shaped area surrounding each cell using a custom MATLAB script42. Cell category-specific activity maps were generated by overlaying a category-specific mask over the activity map of the cognate stimuli (heat for heat-specific and polymodal cells; brush and pinch for mechano-specific cells).
Whole-mount ISH of trigeminal ganglia
Whole-mount ISH of trigeminal ganglia after in vivo imaging and of tissue sections was performed as described previously3 using combinations of the following hybridization chain reaction probes (Molecular Instruments): Trpm8 (GenBank NM_134252; full length), S100b (NM_009115; full length), Fxyd2 (NM_007503; full length), Scn10a (NM_001205321; coding sequence), Calca (NM_007587; full length), Trpv1 (NM_001001445; full length), Tmem233 (NM_001101546; full length), Mrgprd (NM_203490; full length), Nppb (NM_008726; full length), Sst (NM_009215; full length), Mlc1(NM_133241; full length), tagRFP–TVA, tdT and EGFP (which detects GCaMP expression). Two-dimensional dorsal views of the surface of whole-mount ganglia were collapsed by maximum intensity projection from confocal Z stacks with 10-μm intervals to capture the convex surface of the ganglion.
Aligning whole-mount ISH images to in vivo recordings
Alignment of whole-mount ISH images to in vivo fluorescent images was performed as described previously3 using either tagRFP-positive guide-post cells in Tac1–tagRFP/TVA animals or GCaMP-expressing cells stimulated with high K+ directly applied to the trigeminal ganglion. In brief, multi-channel two-dimensional ISH images were crudely aligned to in vivo fluorescence by scaled rotation using the TurboReg plugin and a custom macro in ImageJ/Fiji. Guide-post cells were then manually matched to their in vivo fluorescent counterparts using a custom ImageJ macro that identified coordinate pairs for each guide-post. The ISH image was morphed to match its in vivo counterpart using these coordinates with a custom Python script that builds on the OpenCV library3. Several rounds of ISH were aligned to each other using probes labelling partially overlapping sets of cells in both rounds to provide guide-posts for morphing. As shown previously3, this type of image alignment does not produce a pixel-to-pixel match but accurately identifies ISH-positive cells that respond functionally (Extended Data Fig. 2).
Analysis of gene expression and transcriptomic classification
Cell ROI (responding cells) were manually analysed for expression (negative, weak or strong) of every gene with diagnostic ISH data3 (Extended Data Fig. 2c). Binary expression patterns were decoded into transcriptomic cell classes using the rules outlined in Extended Data Fig. 2b and Supplementary Table 3, which also explains how transcriptomic class nomenclature10 is related to other classification schemes.
Single-cell sequencing data from DRG28 were obtained from GEO Series GSE254789 and analysed with Seurat v.5 in RStudio. Cells with less than 800 expressed genes or more than 5% of mitochondrial transcripts were excluded, and datasets were combined using canonical correlation analysis integration after principal component analysis reduction to 30 components. Neuronal and non-neuronal cell clusters were identified in UMAP by analysing the expression of Snap25, Mbp, Apoe, Qk, Pecam1, Slc17a7 and Slc17a6. Doublets were identified using DoubletFinder v.3, and doublets and non-neuronal cells were removed from the dataset. After neurons were renormalized, reduced to 40 principal components and reintegrated, neuronal clusters were calculated using the Louvain algorithm with a resolution of 0.2 and identified/combined on the basis of the genes shown in Extended Data Fig. 2.
Behavioural assessment of allodynia
For behavioural experiments, groups of adult C57Bl/6, Trpv1–Cre::Avil–LSL-TeNT and control littermates were tested. RNAscope ISH of fresh frozen sections of DRG12 (Advanced Cell Diagnostics) was used to examine the extent and selectivity of TeNT recombination. The cheek or plantar surface of the hind paw was injected with PGE2, LY344864 or phosphate-buffered saline (PBS) as described above. Mice (male and female; more than 8 weeks old) were habituated to the testing chambers for at least two sessions in the days preceding the behavioural tests. When mice were used for more than one experiment, they were allowed at least 7 days to recover between tests. The experimenter was blinded to the genotype of the animals.
For the data shown in Fig. 5a,e, mechanical thresholds (50% withdrawal threshold) were determined using von Frey stimulation by the simplified up–down method59 at multiple time points up to 2 h after injection of PGE2 or LY344864. The experimenter was blinded to the genotypes of the mice. For Fig. 5f, mechanical threshold (60% withdrawal threshold; three responses in five trials) was determined using von Frey stimulation by a standard up–down method at baseline and a single time point 15–30 min after intraperitoneal injection of vehicle (dimethylsulfoxide) or Salvinorin B (Hello Bio; 10 mg ml−1; 10 mg kg−1) and paw injection of LY344864 (as described above). The experimenter was blinded to the injected compound. The mice were tested twice (opposite paws; 6 days apart), with four receiving Salvinorin B in the first test and the other three in the second round. The assignment of mice to the two groups was pseudorandom. Brush allodynia was determined in the same behavioural apparatus by stimulating C57Bl/6 mice. As shown in Extended Data Fig. 9a, 20 brushes were delivered using a paint brush (7950-5 Round; KINGART) once every minute before and 10–30 min after paw injection. A response was counted as any withdrawal from the stimulation. As shown in Extended Data Fig. 9b, a single brush with a fluffed cotton swab was delivered at baseline and at each time point after paw injection of PBS or LY344864. Pain-like behaviours (repetitive or extended lifting and guarding) and brief response to brushing were scored. The experimenter was blinded to the injected compound; thus, mice were randomly assigned to groups.
Spontaneous pain behaviours following the injection of 500 µM PGE2 (in 20 µl of PBS) or PBS alone into the hind paw or cheek were recorded and scored offline using BORIS60 by an observer blinded to the genotype and/or the compounds used. In mice injected in the hind paw, licking of the injected paw was scored for 15 min and quantified for a 10-min period, starting 5 min after injection to match the development of inflammation37. For cheek injection blinding, the animals were randomly assigned to groups. Following injections, the mice were placed in cylindrical plexiglass chambers surrounded by mirrors. All face-directed behaviours and periods of inactivity greater than 1 s were scored for the first 15 min after injection and quantified for the same 10-min period used for the paw. Although inactivity may represent freezing-like behaviour (Supplementary Video 3), it may also represent sitting or sleeping. The single mouse injected with PBS that displayed considerable inactivity did not appear to enter a freeze-like state, whereas the majority of the PGE2-injected mice did. To avoid judgement errors in scoring at the resolution of the videos, inactivity was analysed without trying to assess whether the mouse was in distress. The right cheek was partially depilated 2 days before behavioural recording to aid injection.
Quantification and statistical analysis
The numbers of animals and responding cells that were tested for each transcriptomic class are listed in Supplementary Table 1. All quantification and statistical analyses were performed using Python v.3.8, Pandas v.1.1.3, Numpy v.1.19.2 and Scipy v.1.5.2.
Spontaneous activity was detected as peaks in ΔF/F traces with a minimum prominence of 4% ΔF/F, a minimum absolute peak of 4% ΔF/F and a minimum interpeak interval of 0.6 s using the Scipy find_peaks function. The amplitude of an event was calculated as the difference between peak height and its preceding minimum. Spontaneous activity was quantified over multiple time windows when the cheek or paw was held at 30 °C (105 s for trigeminal neurons and 40 s for DRG) by summing event amplitudes.
Temperature-induced responses were identified as peaks with a minimum prominence of 5% ΔF/F, a minimum interpeak interval of 0.6 s and an onset during the temperature stimulation window. The end of a response was defined as the time point when signal dropped below 10% peak height. The area under the ΔF/F curve from onset to end of the response was used to quantify response to temperature. A cell was considered responsive to a temperature stimulus if AUC exceeded a defined minimum (corresponding to a mean of 3.5% ΔF/F over the 4-s stimulation window). A cell was mechanosensitive if its peak amplitude exceeded 15% ΔF/F during the stimulus application window. Cells were polymodal if the ratio between the mechanical and temperature stimuli was smaller than 5:1 and larger than 1:5.
Quantification of responses for a given stimulus varies according to the choice of cells included in the analysis. For example, cells may have spontaneous activity but not respond to mechanical or thermal stimulation of the cheek. For consistency and to allow comparison between figures, we report the response magnitudes (and numbers) of cheek-innervating neurons, that is, cells that responded to any stimulus applied to the cheek.
To distinguish brush responses from spontaneous activity after chemical induction, individual time-locked brush responses within a 1-s window of stimulus application were identified as peaks with a minimum prominence of 5% ΔF/F and a minimum absolute peak height of 20% ΔF/F. Bona fide brush cells were identified as cells that responded to at least 50% of brushes, which cover largely but not completely overlapping fields of the cheek.
Percentages of transcriptomic cell classes contributing to a functional cell category were calculated as described previously3 by dividing the number of responding cells positive for a given class by the number of responding cells that were tested with ISH probes for that class. Because not all classes were tested in all individual animals, the summed percentages do not necessarily add up to 100%. To display proportions in stacked bar graphs, the percentages were further normalized to 100% in these graphs.
The effects of PGE2 injection were analysed using two-tailed paired Student’s t-test. Attenuation of heat responses by Trpv1 knockout was analysed using one-tailed Welch’s t-test (allowing for unequal variances between different conditions). The effects of Trpv1 knockout on spontaneous activity were analysed using two-tailed Welch’s t-test.
Holm–Šidák correction was applied to all statistical tests to adjust for multiple comparisons when investigating several transcriptomic classes. One-tailed Wilcoxon signed-rank tests were used for von Frey thresholds and brush-induced behaviour when comparing time points after allodynia induction to paired baseline values. In Extended Data Fig. 9b, data were pooled across time points after injection and compared between experimental groups with a chi-squared test. All other comparisons between behavioural groups used the Mann–Whitney U-test. Detailed statistical information is provided in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.