Primary GO annotations
The process for creating GO primary (experimental) annotations from the published literature has been previously described in detail54. New annotations from additional publications are added at the rate of approximately 4,000 per month, and some annotations, if they have been superseded in light of new experimental results or updates in the biological representation captured in the ontology, are revised or removed. Scientific publications used to support experimental GO annotations are labelled with a PubMed LinkOut55 whenever possible and can be retrieved at https://pubmed.ncbi.nlm.nih.gov/?term=loprovGeneOntol%5bSB%5d. A small number of additional publications are not indexed by PubMed. Our analyses used the ontology and annotations from the GO knowledgebase release 22-03-2022 (https://release.geneontology.org/2022-03-22/index.html, https://doi.org/10.5281/zenodo.6399963). There were 713,330 primary annotations, including 147,872 annotations to human genes and 565,458 to genes in other organisms. For all annotation counts, we excluded direct annotations to the class ‘protein binding’, as these statements represent observed interactions but are not descriptions of function in the same sense as other GO annotations56, and are therefore not considered for inclusion in the PAN-GO set.
Overview of the evolutionary modelling approach
Our approach13 brings together all experimentally supported GO annotations for all members of a gene family, in the context of a phylogenetic tree representing how those genes are related, to generate a model of the evolutionary process by which the members obtained the functions they now possess. This is a longstanding, standard method for reconstructing the evolution of traits or characters that is commonly applied to species28,29,30,31. Here we applied a similar approach to trees of genes rather than species and to functional characteristics rather than phenotypic characters. However, modelling gene functional characteristics involved the major additional challenge that the experimental data are sparse and highly unevenly distributed. Genes have been studied to widely varying degrees depending on scientific and medical interest, and this interest has been largely concentrated on genes in humans and a handful of model organisms. To address this challenge, we also use many other pieces of evidence, such as protein domain structure, known active-site residues, free-text function descriptions from the UniProtKB/Swiss-Prot knowledgebase37, among others.
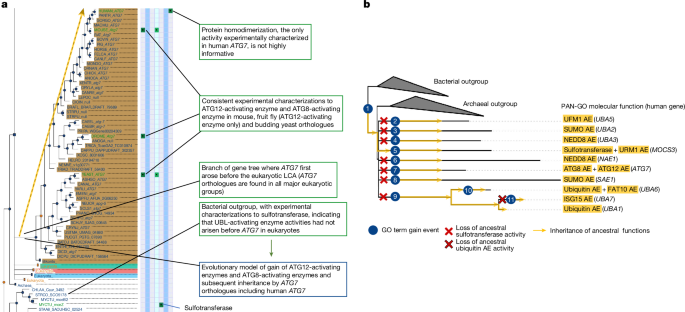
For each gene family, we generated an evolutionary model that specifies how each functional characteristic, represented by a GO class, was gained or lost during evolution. Specifically, we describe the evolution of function in terms of three types of event: root, gain and loss. A root event is defined as a GO class that is inferred to have been present in the LCA of the protein family. A gain event is defined as a GO class that was not (or cannot be confidently inferred to be) present in the LCA of the entire family, but arose later along a specific branch of the tree. A loss event is defined as a GO class that had arisen earlier (through a root or gain event) but was subsequently lost along a specific subbranch of the tree (that is, in some but not all descendants of the original root or gain).
Every root or gain event must be supported by direct experimental evidence in at least one, but often multiple, of the descendants of the root or selected branch of the tree. As a result, each event is based on a combination of traceable experimental evidence and curator inference of the point in evolution (the root or a specific branch in the tree) at which this function first appeared. The Evidence and Conclusion Ontology (ECO)57 evidence code IBD (ECO:0000319 ‘inferred from biological descendant’) was used to denote this type of evidence, and all genes with experimental evidence are stored as metadata to provide a traceable evidence trail. Loss events prevent GO classes from being inherited by specific subclades that descend from a gain event; the evidence used for loss events is described in more detail below.
The evolutionary model for the family was then used to create inferred annotations for each family member based on inheritance from ancestors in the tree: a GO class is inherited by all children of a root or gain event for that class unless a loss of that same class is encountered along the path in the tree. All family members will therefore receive the same GO annotations if the family has only root events, but different annotations if there are any gain or loss events along specific internal branches of the tree. These inferred annotations comprise the set of human gene functions we describe here and can be identified in the GO knowledgebase by the ECO code ‘inferred from biological ancestor’ (IBA) (ECO:0000318). Each IBA annotation also includes the following metadata for providing a traceable evidence trail: (1) the persistent identifier of the tree node from which the annotation was inherited (the root node or terminal node of the annotated gain branch); and (2) the source of the experimental data used to support the root or gain event.
PAN-GO evolutionary modelling process
A more detailed description of the process of producing and updating PAN-GO annotations is shown in Extended Data Fig. 4. The process includes manual construction of an evolutionary model for each family, using as input PANTHER phylogenetic trees and primary GO annotations. Both automated and manual updates are performed in response to user feedback, changes in biological knowledge in the ontology, changes in primary annotations and changes in PANTHER tree topology. Updated PAN-GO gene annotations (IBA) are generated monthly from these updated models. The different steps leading to the final PAN-GO gene annotations are described in this section.
Phylogenetic trees
The gene trees were obtained from the PANTHER knowledgebase15. The PAN-GO annotation set presented here was generated using v.15.0 of the knowledgebase, released in 2020. Trees were constructed using the GIGA tree reconstruction algorithm58 for protein-coding genes in 142 organisms that span the tree of life, but the selection of organisms (https://pantherdb.org/panther/speciesTree.jsp) was biased with the aim of reconstructing genome evolution in humans and well-studied model organisms. The trees were fully reconciled with the known species tree, and all nodes were annotated by event type (speciation, gene duplication and horizontal gene transfer) and the common ancestor species or clade for speciation nodes. Each tree has an associated protein sequence alignment that was used to reconstruct the phylogeny. Protein sequences were obtained from the UniProt Reference Proteomes resource37, which selects one canonical protein sequence per protein coding gene in each genome.
Creating curated models of function evolution
To implement the PAN-GO process, we created a specific software tool for manual curation of function evolution models, which we call PAINT13. The PAINT user interface provides an integrated view of the phylogenetic tree, a matrix of experimental GO annotations structured by ontology relationships, a multiple sequence alignment annotated with functional sites from UniProt/Swiss-Prot records37 and domains from the Pfam resource5. It also displays brief free-text descriptions of the protein products of each gene in the tree, protein names and links to pages in knowledgebases including UniProt/Swiss-Prot and model organism databases. PAINT enables expert biocuration scientists to transform the input information, a phylogenetic tree with experimental GO annotations on terminal (leaf) nodes of the tree, into an output evolutionary model as described above. The specific guidelines for constructing models of function evolution in a protein family, to promote consistency and reproducibility of the evolutionary models, are detailed at https://wiki.geneontology.org/PAINT_User_Guide. Curators also meet regularly to review sample families from each curator, to review and to cross-check the evolutionary models. The evolutionary models are saved to a relational database and can be accessed and viewed at https://pantree.org. The PAN-GO annotations derived from the models are exported in Gene Annotation Format (GAF) (https://geneontology.org/docs/go-annotation-file-gaf-format-2.2/) and deposited in the GO knowledgebase. They are also included in the data distributed by providers of GO annotations such as UniProt-GOA59. These annotations are labelled with the evidence code IBA, and contain metadata with details of the evidence or provenance, including the curated tree node from which it inherited its function (represented as a stable PANTHER tree node identifier) and the genes providing the original experimental evidence. Source code for the PAINT tool is available at GitHub (https://github.com/pantherdb/db-PAINT).
Inspection of the phylogenetic trees
The first step of the PAN-GO curation process consists of the analysis of the structure of the phylogenetic tree to gather clues about the evolution of the family. Speciation, duplication and horizontal transfer events are closely considered. Speciation events define the age of the family and the taxonomic distribution of related genes in different clades. This information helps guide the choice of GO classes based on the functions known to occur in the species present in a tree or subtree. A more ancient ancestor (which generally leads to a wider species distribution) may lead to more conservative annotations owing to uncertainty in reconstructing ancient functions. The tree can also provide other important clues for identifying functional evolution events. Duplication events are examined closely as these events often lead to gain and/or loss of functions. Horizontal gene transfers, which include some eukaryotic mitochondrial or plastid genes with origins in ancestral prokaryotic endosymbionts, are also carefully evaluated, as functional characteristics of a transferred gene may have been modified after transfer.
Application of taxonomic restrictions
Because of the high diversity of living organisms, it is not possible to cover all species with a taxon-neutral ontology and there are inherent taxon specificities in many branches of the GO ontology. An iconic example is the cellular component ‘mitochondrion’, which is specific to eukaryotes. Explicit formalization of taxon constraints60 are used to avoid taxon-inappropriate annotations. The PAINT curation tool highlights any inconsistencies between taxon constraints and annotations when constructing an evolutionary model.
Analysis of the experimental evidence
The analysis of all the experimental data available enables the selection of the most relevant classes that will be used in the evolutionary model for a gene family. An essential indicator is the consistency of the MF, BP and CC classes associated with the various members across species represented in the tree. If the annotations in a clade of related genes are consistent, they are likely to have all inherited those aspects of function from their LCA, which suggests that those functions evolved before the LCA. If they are inconsistent, a curator attempts to identify consistent subclades that evolved a different function, or gained or lost a function. Assessing consistency among GO classes that are not explicitly related in the ontology structure is challenging and often requires deep biological knowledge on the part of the curator. To decide which classes are appropriate to be associated with members of a protein family, the PAN-GO curators use additional sources: they can review the content of model organism databases or UniProtKB/Swiss-Prot (https://www.uniprot.org) through direct links provided by the PAINT tool. Curators often assess additional references to confirm or invalidate certain data. Finally, the presence of particular predicted sites and domains (active sites, transmembrane regions or protein domains) may lend more support for specific functions having evolved along particular branches in the tree.
Selection of the most informative annotations
In principle, PAN-GO curation could have resulted in an evolutionary root or gain event in the tree for every GO class that was annotated to at least one family member from experimental evidence. In practice, however, there is often considerable redundancy and overlap between these GO classes, and not all terms represent actually distinct functional characteristics. Consequently, the PAN-GO curation process is selective. We provide some examples below. To provide a quantitative estimate of the selectivity, we counted, for each family, the number of nonredundant function classes (that is, excluding annotations to more general classes in the ontology) that were available to a curator; these were all the classes that could have been used in the evolutionary model for the family. We then calculated the number of classes actually used in the evolutionary model for each aspect of the ontology. Extended Data Table 2 shows the average of these values over all families. On average, only 24%, 28% and 13% of the experimentally annotated MF, CC and BP GO classes, respectively, were annotated to root or gain events during the phylogenetic curation process. In general, this high selectivity is due to the integrative aspect of the process: all experimental GO annotations for all family members are considered as a whole. By contrast, an experimental GO annotation is designed to capture a specific finding from experiments reported in a single publication. As a result, a PAN-GO curator can select the most informative GO classes among the experimental annotations and recognize when different experimental annotations are likely related to the same underlying function. Often, functionally related terms are also related in the ontology (the PAINT tool groups together hierarchically related terms to facilitate the selection process). Curators can then distinguish such apparent functional differences from actual functional differences among family members. Extended Data Table 2 shows that the PAN-GO curation process results in selection of a relatively small fraction of GO biological process classes compared with the other aspects of the GO ontology. This is due in part to the complexity of the biological process branch of the ontology (around 30,000 classes versus <10,000 each for MF and CC), and partly due to less stringent criteria for involvement in a process versus the other aspects. Many of the excluded classes are either related but less informative classes or downstream effects of the primary functions of the gene, such as peripheral functions or phenotypes and readouts that represent consequences of a gene’s function but not accurate descriptions of the function itself.
An example of BP class selection is shown in Extended Data Fig. 5a: the regulation of production of various interleukins and transcription of downstream targets are observations (experimental readouts) for the ‘cytoplasmic pattern recognition receptor signalling pathway’. There are several reasons that primary annotations for the same underlying function often use related, but not identical, GO classes: primary annotation is spread out in both space and time, and each species is often treated by a different curator. This is compounded by the fact that some functional characteristics (GO terms) are important in a few species but too specific for inclusion in the evolutionary model. Moreover, the authors of the articles from which the primary annotations are drawn use widely varying terminology. Primary GO annotations that are only supported by data from large-scale experiments (most typically, cellular localization) or annotations inconsistent with all other data available for the family are set aside until there is strong support by other annotations.
In many cases, parent and child classes (indicating less specific, representations of a functional characteristic) are both used for primary annotation throughout the families, but only the most relevant ones are selected in the PAN-GO process (Extended Data Fig. 5b): the GO terms ‘regulation of innate immune response’ and ‘cellular response to virus’ are more general classes for the concept ‘antiviral innate immune response’, which is more representative of the function of genes in the family. It is the integrated analysis of the family and its primary annotation that enabled the PAN-GO curator to select the most appropriate class (or classes) to include in the evolutionary model.
As in the ‘three blind men and the elephant’ parable, primary annotations, which describe individual experimental observations, are generally correct but sometimes only tell part of the story. The goal of the PAN-GO curation is to provide a more integrated picture whenever possible while still providing a comprehensive set of GO function annotations.
Capturing loss of function and preventing inheritance of low-confidence annotations
Loss of function is based on specific types of evidence when available. In some cases, negative primary GO annotations (indicated by the NOT qualifier) are available, and in this case the loss event (like root and gain events) uses the IBD evidence code. In other cases, when important residues or domains are known to be required for the function, multiple sequence alignments can reveal the absence of these important features in some branches and provide evidence for the loss of function; these are denoted with the ‘inferred from known residues’ (IKR) (ECO:0000320) evidence code. The loss of function due to mutations in specific amino acids such as active site residues is well characterized in the literature for some families (for example, PTHR24418, non-receptor protein kinase family). For families with relatively well-studied genes, it is often possible to infer that a lack of corroborating GO annotations suggests that the function has been lost; in these cases, curators check the UniProtKB/Swiss-Prot knowledgebase as well as the literature to increase the confidence of such inferences. In less well-studied families (that is, with sparse experimental GO annotations), curators may decide to introduce a loss (particularly after gene duplication) to avoid false-positive annotations. These events are denoted by the ‘inferred from rapid divergence’ (IRD) (ECO:0000321) evidence code. The main purpose of this step is to remain conservative in the PAN-GO inference process to ensure the high quality of the annotation set produced. It should be noted that loss events labelled with IBD or IKR result in negative GO annotations (indicating that a gene does not possess a given functional characteristic), and these annotations are available in the GO knowledgebase. However, for clarity, we do not include negative annotations in the PAN-GO set of human gene functions available at https://functionome.geneontology.org, and these appear only in the evolutionary models.
Annotations for genes that were not in a PANTHER family
There are 994 human genes that are not currently in a PANTHER family, and these mainly encode short proteins, many of which do not exhibit clear evolutionary conservation. Only 114 of these genes had primary annotations. For 61 of these genes, we were able to select informative primary annotations and included them in the PAN-GO set of human gene functions.
Staying current with evolving knowledge
As the GO ontology and primary gene annotations are constantly being expanded and revised in response to new experimental data and interpretation, the PAN-GO process includes an automated updating and publishing step after each new GO knowledgebase release (approximately monthly) or each new PANTHER release (yearly). In addition, issues identified by feedback from GO curators and the wider GO user community lead to manual review of the ancestral annotations (or, much less commonly, trees) as appropriate. The PAN-GO project has developed an extensive software suite to support these updates and improvements.
Addressing changes to GO classes and annotations
The monthly automated updating step after each new GO knowledgebase release handles any required action due to changes in the ontology classes (terms) or experimental GO annotations that were used as evidence for the functional evolution events in the evolutionary model. These actions include updates for obsolete and merged classes, and the removal of any annotation no longer supported by experimental data or failing taxon restrictions.
Evolutionary models are also updated according to the availability of new experimental data and subsequent primary GO annotations, as new classes and new annotations cannot be integrated automatically but go through manual analysis of the experimental evidence. For instance, during the complete review of the ontology associated with transcription, the class ‘histone chaperone activity’ was created, and primary annotations were revised. This new class was used to update the evolutionary models of applicable PANTHER families such as PTHR21315 or PTHR12040.
Addressing updates to the topology of phylogenetic trees
The phylogenetic trees are updated after release of new PANTHER versions, based on the annual release of the protein sequence data from the UniProt Reference Proteomes and Quest for Orthologs efforts61. PAN-GO evolutionary models refer directly to stable tree-node identifiers; that is, each gain and loss event is associated with the identifier for the terminal node of the branch along which the event occurred. As tree-node identifiers are retained between PANTHER versions whenever possible, the PAN-GO annotations for those branches are retained in the newer version of PANTHER trees. However, improvements in tree reconstruction algorithms and the addition of more species sometimes lead to modifications of the family structure: some families can be split into several smaller families or merged into a single, larger family. Consequently, some branches can move from one family to another or be lost. When this happens to a branch that was annotated in a PAN-GO evolutionary model, a ‘require review’ notification is added to the affected families, and curators review and revise the evolutionary models when necessary.
Addressing user feedback
Extensive feedback from experts from several model organism databases permitted the addition of an extra layer of quality control to the PAN-GO evolutionary models. Feedback is handled through the GO annotation issue tracker in GitHub (https://github.com/geneontology/go-annotation/labels/PAINT%20annotation). The two largest contributors of feedback tickets have been PomBase, the scientific resource for Schizosaccharomyces pombe (fission yeast) (https://www.pombase.org/)62, with nearly 600 update requests, and FlyBase, the scientific resource for Drosophila melanogaster (fruit fly) (https://flybase.org/)63, with over 200 update requests, over a 7 year period. The genomes of Drosophila species contain many traces of more or less ancient duplication events, which also enable a better understanding of these events in the whole phylogenetic tree and contribute to improving our evolutionary models of gain or loss of functions64. The other resources in the GO Consortium, including model organism databases and UniProtKB, also contributed to the validation of the annotations (total of 100 update requests).
Analysis methods
Accessing and using the human PAN-GO annotations
The PAN-GO annotations used for the analyses presented here can be downloaded at https://functionome.geneontology.org/download/functionome_release.gaf.gz.
Estimating the reliability of PAN-GO annotations
There is no absolute source of truth that enabled us to assess the correctness of GO annotations. To address this problem, a surrogate measure called ‘reliability’, which can be calculated for GO annotations, has been previously proposed38. This measure takes advantage of the fact that GO annotations are being added and removed over time, and they can be compared at different time points to calculate the reliability of older annotations. Specifically, if an experimental annotation is later added to the GO knowledgebase that is to the same or more specific term than an older annotation, the older annotation is considered to be confirmed. Conversely, if an experimental annotation is later added to the GO knowledgebase that uses the NOT qualifier (indicating that a gene has been shown NOT to have that functional characteristic) and is either the same or less specific than the older annotation, the older annotation is considered to be rejected. Because NOT annotations are rare in the GO knowledgebase, the number of rejected annotations is low in practice, thereby leading to an inflated reliability. The previous study38 suggested that another property could be calculated, the number of older annotations that were later removed, based on the assumption that they were later judged to be incorrect. They then defined reliability as:
$${\rm{Reliability}}={N}_{{\rm{confirmed}}}/({N}_{{\rm{confirmed}}}+{N}_{{\rm{rejected}}}+{N}_{{\rm{removed}}})$$
(1)
where Nconfirmed is the number of GO annotations present in an older version (at time point t0) of an annotation set, which were later confirmed before time point t1, Nrejected is the number of GO annotations present at time t0 that were rejected between time points t0 and t1, and Nremoved is the number that were removed between time t0 and t1.
Using this method, we calculated the reliability of PAN-GO annotations. We first gathered all primary annotations made between October 2019 and March 2022 from the GO knowledgebase using the date stamp on each annotation. We then compared them with the PAN-GO annotations in the October 2019 release of the GO knowledgebase. The comparison included 11,102 new primary annotations and 21,145 PAN-GO annotations for the same set of 4,007 human genes. If the GO class from the new primary annotation is the same or more specific as that of a PAN-GO annotation, the PAN-GO annotation is considered to be confirmed. By this definition, 2,354 PAN-GO annotations for 1,608 genes were confirmed. Extended Data Table 3 shows the breakdown of the confirming primary annotations by evidence code; most of these derive from direct assays on a specific gene product (IDA), and only 29 were from high-throughput studies (HDA).
Of the new experimental annotations, there were 54 negative (NOT qualifier) annotations, of which only three disagreed with PAN-GO annotations. After reviewing these three negative annotations, we found that one was specific to one protein isoform but not the canonical protein encoded by the gene (so the PAN-GO annotation is correct), and the remaining two were to the same transporter gene and refer to zinc as a substrate (SLC30A10 NOT ‘zinc ion transmembrane transporter activity’, and SLC30A10 NOT ‘intracellular zinc ion homeostasis’). However, other papers (supporting other primary GO annotations) have demonstrated these same functions for SLC30A10, and therefore confirm the PAN-GO annotations. As a result, there were 0 negative GO annotations that can be considered to reject PAN-GO annotations. We recognize that 54 negative annotations is a small sample, which will underestimate the actual PAN-GO error rate. Following the previously described method38, we also examined the PAN-GO annotations that were present in our October 2019 release but later removed. We found 4,809 PAN-GO annotations had been removed, but in most cases, annotations were removed owing to redundancy with another, more informative PAN-GO annotation (fine-tuning of the annotation set) and not because of an error. To estimate an error rate, we reviewed a random sample of 500 removed annotations and categorized each one as correct but not meeting PAN-GO selection criteria (fine-tuning of selected annotations for modelling), incorrect (selection in the evolutionary model of an experimental annotation that is actually incorrect) or uncertain (demonstrated in a homologue but possibly incorrect for the annotated human gene). We found that 7 (1.4%) were incorrect and 20 (4%) were uncertain. Assuming these percentages approximately hold for the entire set of removed annotations, we estimated that between 67 (removed because they were incorrect, 4,809 × 1.4%) and 260 (removed because they were either incorrect or uncertain, 4,809 × 5.4%) were removed because of errors. This would give a reliability (equation (1) above) of PAN-GO annotations between 90% (260/(2,354 + 260)) and 97% (67/(2,354 + 67)).
One example of a clearly incorrect PAN-GO annotation was found within the carnitine O-acyltransferase family (PTHR22589). CPT1C, in contrast to the CPT1A and CPT1B paralogues, does not have ‘carnitine O-palmitoyltransferase activity’ in mitochondria, but localizes in the endoplasmic reticulum where it shows ‘palmitoyl-(protein) hydrolase activity’65,66. This type of incorrect inference of functional conservation through ancient duplication events, and therefore errors in evolutionary modelling, is one of the most common errors we found during our review. When such errors are discovered, the PAN-GO evolutionary model is updated to correct the error.
A relatively frequent case of important fine-tuning of PAN-GO annotations relates to the sometimes subtle difference between a GO term for a BP and the corresponding GO term for regulation of that process. Frequently, the primary annotation derived from an experiment, often based on the effects of a genetic manipulation such as a deletion, uses the regulatory term. Other experiments, however, may show that the protein in question is directly involved in the process (resulting in an annotation to the process itself rather than its regulation). Several PAN-GO annotations were updated (5 in our sample of 500) to consistently reflect either involvement in, versus regulation of, a particular BP. Other common updates were due to inconsistencies in the primary annotations for enzyme complexes to the GO term ‘complex assembly’ (10 in our random sample of 500), which we consider to be fine-tuning as they are correct even if not highly informative.
Broad functional categories on the PAN-GO website
To facilitate browsing of the PAN-GO annotations, and for visualizing the landscape of human gene functions in Fig. 3, we mapped each annotation to a set of selected, relatively high-level GO categories. Broad functional categories were taken from the generic GO subset, which is available at https://release.geneontology.org/2022-07-01/ontology/subsets/goslim_generic.obo. Note that these are categories of annotations, not genes, so a gene annotated to multiple distinct GO terms may appear in multiple categories. Note also that some of these broad categories are subcategories of others; in this case, a gene was assigned only to the more specific subcategory, and not the more general category, to minimize the overlap between categories and therefore facilitate visualization and browsing.
PAN-GO annotation browser
We developed a simple web-based tool for exploring the set of human gene functions, including links to all experimental evidence and phylogenetic trees. It is implemented using ElasticSearch and is available at https://functionome.geneontology.org/. Code is available from GitHub (https://github.com/pantherdb/pango).
Contributions of experimental evidence from model organism annotations
Primary GO annotations (supported by published experimental evidence) are used for all PAN-GO annotations. We characterized this evidence in detail for each model organism (Extended Data Table 1). Column 2 reports the number of PAN-GO annotations that are supported by one or more publications with experimental evidence for function of a gene in that organism. Evidence obtained from experiments on human genes is divided into two rows: one for direct evidence for a given gene and one for evidence for a related (paralogous) human gene. Column 3 reports the number of PAN-GO annotations supported only by experimental evidence for homologous genes (that is, it excludes any PAN-GO annotations that have direct experimental evidence for the human gene). These annotations were inferred from other human paralogues or non-human homologues, but have not yet been experimentally confirmed. Column 4 counts PAN-GO annotations that are based on non-human experimental data only. Column 5 counts PAN-GO annotations that are based on evidence from only one species. Column 6 counts all experimental annotations in each organism that could potentially be used as literature evidence for human PAN-GO annotations.
Evolution of gene functions
For each PAN-GO annotation, we retrieved the branch of the evolutionary tree that was modelled as having gained that functional characteristic, representing when that characteristic first evolved in an ancestor of a human gene. Because the phylogenetic approach defines ancestors in terms of LCAs of extant species, our evolutionary model specifies the interval between two of these LCAs, during which the functional characteristic evolved. The approximate dates for each of these LCAs has been determined67, so we could convert the LCA interval to a time interval. For instance, if a gene function characteristic now found in a human gene first appeared along the branch leading from the LCA of Eukaryota and Archaea (around 4,250 million years ago) to the LCA of plants and animals (the LCA of Eukaryota, about 1,598 million years ago), then the function first evolved between 4,250 and 1,598 million years ago, and was then transmitted unchanged from parent to child for at least 1.6 billion years all the way to modern humans.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.