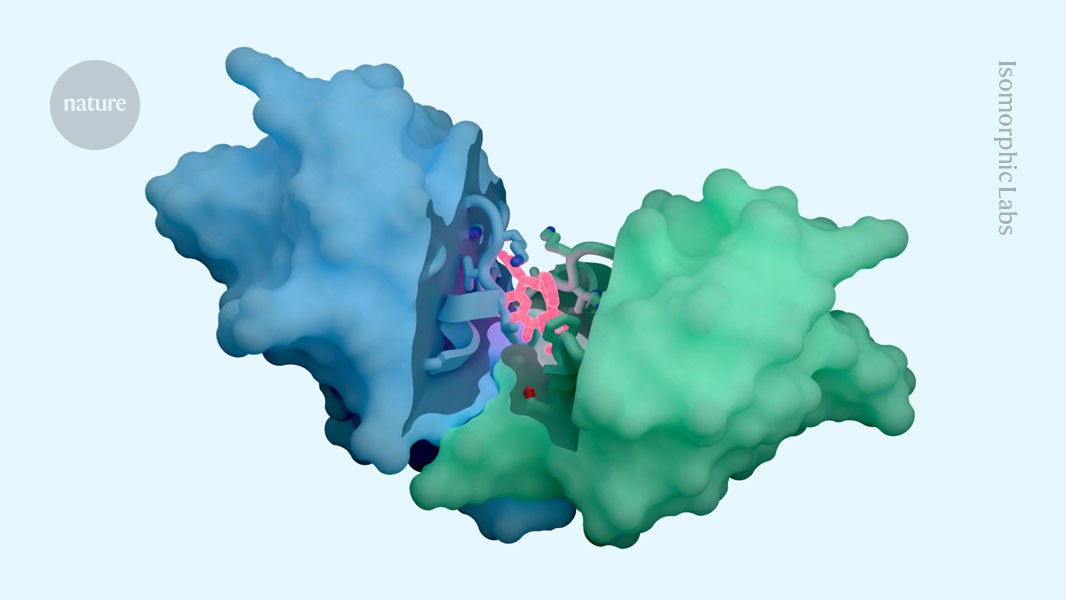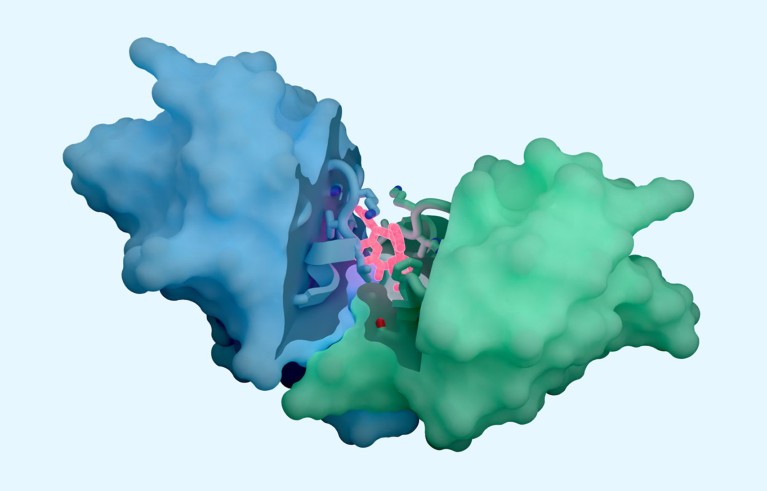
The AI tool includes predictions of how proteins interact with potential therapeutic molecules.Credit: Isomorphic Labs
Nearly two years after Google DeepMind released an updated AlphaFold3 geared at drug discovery, its biopharmaceuticals spin-off, Isomorphic Labs, announced an even more powerful artificial-intelligence model — and they’re keeping it all to themselves.
Isomorphic Labs, based in London, touted the capacities of its ‘drug-discovery engine’ — which it calls IsoDDE — in a 27-page technical report, released on 10 February. Achievements, including precise predictions of how proteins interact with potential drugs and antibody structures, have impressed scientists working in the field.
Yet unlike the AlphaFold AI systems for predicting protein structure — which were made accessible to other researchers and described in depth in journal articles1,2 — IsoDDE is proprietary, and the technical paper offers scant insight into how to achieve similar results.
AlphaFold is five years old — these charts show how it revolutionized science
“It’s a major advance, on the scale of an AlphaFold4,” referring to an unreleased future generation of Google DeepMind’s technology,says Mohammed AlQuraishi, a computational biologist at Columbia University in New York City who is working to develop fully open-source versions of AlphaFold. “The problem, of course, is that we know nothing of the details.”
Drug–protein interactions
AlphaFold 3 was developed with drug discovery in mind. Unlike its Noble-prizewinning predecessor AlphaFold2, the model could predict the structures of proteins interacting with other molecules — including potential drugs.
Similar AIs modelled after AlphaFold 3 have come close to fully matching its performance and have new capabilities. An open-source model called Boltz-2, developed by scientists at the Massachusetts Institute of Technology in Cambridge and released last year3, could predict the strength to which potential drugs glom onto proteins, or binding affinity. This is a key property for developing therapeutics and is usually predicted with computationally intensive physics-based methods.
AlphaFold is running out of data — so drug firms are building their own version
According to Isomorphic’s report, its new AI outperforms both Boltz-2 and physics-based methods at determining binding affinity. Predictions of how antibodies — which form the basis for therapies that rack up tens of billions of pounds in sales annually — interact with their targets is also state of the art, the report claims.
AlQuraishi says he is especially impressed by the IsoDDE’s ability to predict drug–protein interactions of molecules that are vastly different from the data that the model was trained on. “That’s the really hard problem, and suggests that they must’ve done something pretty novel,” he says.