Genetically modified mice
All experimental protocols and animal care and handling were approved by the Ethics Committee of Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (XHEC-F-2024-086). The bicistronic expression vector px330-Cas9 (Addgene, 42230) containing the Cas9 coding region and the sg sequence (5′-ACATATGCTTCGGAGACTCA-3′) was directly used for microinjection. A 134 bp oligonucleotide acted as the donor ssDNA for intracytoplasmic DNA microinjection (134 bp oligonucleotide sequence, with the edited bases indicated in bold: 5′-GAGTTTGCCGACATAT CCAAAGAGGACCAGATTAAGAAA CTGCATGATTTGCTGGGGCCACACATGCTGTGGAGACTCAAGGCAGATGTCTTTAAGAACATGCCAGCCAAGACCGA GCTCATTGTTCGAGTGGA-3′). For zygote intracytoplasmic DNA microinjection, the mixture of px330-Cas9-sg plasmid (50 ng μl–1) and 134 bp ssDNA donor (100 ng μl–1) was diluted in RNAase-free water.
All mice were bred and maintained according to Shanghai Laboratory Animal Center Institutional Animal Regulations. Mice were housed in a specific pathogen-free animal facility at the Center for Excellence in Molecular Cell Science (CEMCS) with autoclaved food, bedding and water. Animals were maintained at room temperature (20–25 °C) at a humidity of 30–70% on a 12/12-h light–dark cycle (7:00–19:00). All mouse studies were carried out following the guidelines of the Institutional Animal Care and Use Committee (IACUC) at the CEMCS. B6D2F1 mice (female C57BL/6J × male DBA/2) aged 6–8 weeks were used for zygote collection. ICR mice, purchased from Beijing Vital River Laboratory Animal Technology, were used for pseudo-pregnant foster mother and vasectomized males. Mice were generated through standard mouse breeding procedures in the CEMCS animal facility The Chd3emh34 (endonuclease-mediated mutation-human 34 bp) mouse line was generated through homologous recombination using CRISPR–Cas9 technology. For the Chd3emh34 mouse line, a 34 bp mouse sequence (5′-ACTGGGGCCACATATGCTTCGGAGACTCAAGGCG-3′) was exchanged for the human sequence (5′-GCTGGGGCCACACATGCTGTGGAGACTCAAGGCA-3′); edited bases are highlighted in bold.
B6D2F1 female mice (8 weeks old) were injected with 7 IU (international units) of pregnant mare’s serum gonadotropin (PMSG). Subsequently, 7 IU of human chorionic gonadotropin (hCG) was injected over 46-48 h. Female mice were housed with B6D2F1 male mice overnight. Zygotes were collected from oviducts of B6D2F1 females 24 h after hCG injection using hyaluronidase (Sigma, H3884). The px330-Cas9-sg plasmid and 134 bp ssDNA donor was thoroughly mixed before injection. Using a micromanipulator (Olympus) and a FemtoJet microinjector (Eppendorf), the injection mixture was injected into zygotes. The injected zygotes were cultured in AA-KSOM (Millipore, MR-106-D) medium for 24 h in an incubator at 37 °C with 5% CO2 until they developed into two-cell embryos.
The pseudo-pregnant foster mothers were prepared by mating oestrous ICR female mice with vasectomized male mice on the same day as the injection. The two-cell embryos were transferred into oviducts of 0.5 days post coitum (d.p.c.) recipient. Recipient mothers delivered pups at 19.5 d.p.c.
The neonatal mouse tissues were lysed with lysis buffer (100 mM Tris HCl (pH 7.8), 0.2% SDS, 5 mM EDTA, 200 mM NaCl and 100 μg ml–1 proteinase K) at 55 °C for more than 6 h, and then the mixture was boiled at 95 °C for 10 min to deactivate proteinase K. All mice were genotyped with specific primers (primer-F: 5′-TTAGCAACTTGGAGGGCTTC-3′; primer-R: 5′-TCTGCATGGGGCCTAGCTCC-3′). The Chd3R1025W mutation was verified by Sanger sequencing. The primer sequences used for mouse construction and genotyping are listed in Supplementary Table 7.
Primary mouse cortical neuron culture
Mouse cortical neurons were extracted from 14.5-day-old embryos of either sex. Cerebral cortices were dissected and digested with 20 U ml–1 papain (LS003126, Worthington) at 37 °C for 30 min, then cultured at 100,000 cells per cm2 on Lab-Tek II chamber slides (154941, Thermo Fisher Scientific) in 0.5 ml per well of Neurobasal medium (21103-049, Gibco) supplemented with 0.2% B27 (17504-044, Gibco) and 2 mM GlutaMAX (35050-061, Gibco). Lipid transfection using Lipofectamine 2000 (11668-019, Invitrogen) with 0.2 μg of each vector was performed after 24 h of cultivation following the manufacturer’s protocol. The medium was changed every 2 days. For morphological analyses, neurons were immunofluorescently stained at day 3 in vitro to assess axonal morphology and at day 7 in vitro to evaluate dendritic morphology. Confocal microscopy was used to acquire images, and the total axonal length, axonal neurite length, dendritic branch numbers and dendritic branch length were quantified using the Simple Neurite Tracer plugin in ImageJ. The mouse Chd3 shRNA sequence is presented in Supplementary Table 8.
HEK293T cell culture and preparation of protein samples
HEK293T cells were cultured to approximately 90% confluency in a culture dish (Corning) with DMEM (11965092, Gibco) and 10% FBS. Cells were transfected with the corresponding expression plasmids using Lipofectamine 2000 (11668-019, Invitrogen) according to the manufacturer’s instructions. After 72 h of incubation, the cells were washed with 1× PBS, collected via gentle pipetting and lysed in loading buffer. The lysates were heated at 100 °C for at least 30 min to ensure complete protein denaturation and stored at −20 °C. Proteins from cultured neuronal cells were extracted using the same protocol as for HEK293T cells.
RNA extraction and reverse transcription
Total RNA was extracted from cultured mouse cortical neurons or HEK293T cells using TRIzol reagent (15596018, Invitrogen) following the manufacturer’s instructions. Reverse transcription was performed with PrimeScript RT master mix (RR036A, Takara Bio) according to the provided protocol, using 500–1,000 ng total RNA as the template for each PCR reaction.
qPCR
qPCR was performed to measure the relative mRNA expression of Chd3, normalized to Gapdh. The experiments were conducted using SYBR Green PCR Premix (QPK-201, TOYOBO), and data were analysed on a StepOnePlus Real-Time PCR system (Applied Biosystems). The qPCR program included an initial denaturation at 95 °C for 10 min, followed by 40 cycles of amplification at 95 °C for 10 s, 60 °C for 15 s and 72 °C for 20 s. Relative mRNA levels of Chd3 were calculated and normalized using the ΔCT method, with Gapdh serving as the internal control. The primers used are listed in Supplementary Table 9.
Immunoblotting
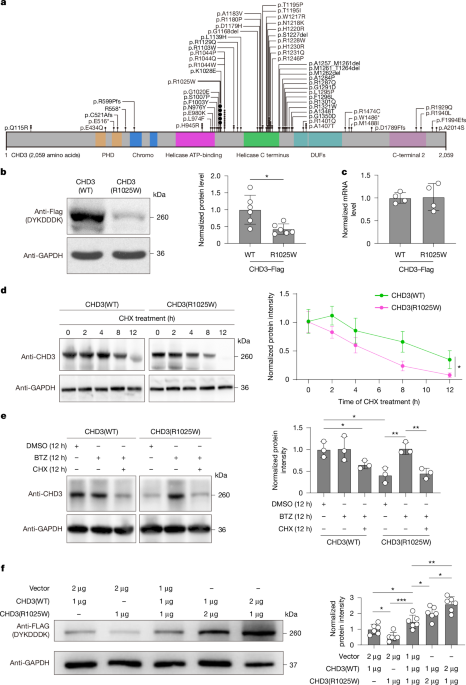
Protein samples from mouse tissues were lysed using radioimmunoprecipitation assay (RIPA) buffer containing 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl (pH 7.4), 1% Triton X-100 and protease inhibitor cocktail tablets (04693159001, Roche). The tissue samples were thoroughly digested on a rotating shaker at 4 °C, and the lysates were centrifuged at 10,000g for 10 min at 4 °C. The resulting supernatant was collected as the protein sample and heated at 100 °C for at least 30 min to ensure complete denaturation. The samples were then subjected to gel electrophoresis using SDS–polyacrylamide gels (Precast gel Tris-Gly 4–20%, 10 wells, 1.5 mm, GSG2001-420T) at a constant voltage of 80 V for stacking and 120 V for separation. Proteins were transferred onto polyvinylidene fluoride membranes (pore size of 0.45 μm, Merck Millipore) at a constant current of 200 mA. Membranes were blocked with 5% bovine serum albumin (BSA) diluted in 1× Tris-buffered saline (TBS)-Tween 20 for 2 h at room temperature, followed by overnight incubation at 4 °C with primary antibodies. After washing with 1× TBS-Tween 20, membranes were incubated with secondary antibodies for 1 h at room temperature. Protein bands were visualized using chemiluminescence (ECL ImmunoBlotting Substrate, 32106, Pierce). Raw images of the immunoblots are shown in Supplementary Fig. 1.
AAV plasmid construction and production
The plasmid vectors related to AAV virus packaging were constructed as previously reported29. In brief, we generated the AAV vector through golden gate assembly and restriction enzyme digestion, the first part of which contains the human U6 promoter to initiate the expression of sgRNA and the human synapsin promoter to express split spCas9 protein (amino acids 1–1029) conjugated to the intein-N element. The second part of the AAV vector includes the human synapsin promoter to trigger the expression of split spCas9 protein (amino acids 1030–1368), in which the TadA*(LOF)-TadA*(F148A) segment is embedded at the 1249 site and replaces the amino acid 1249–1263 segment of spCas9. Note that the WPRE3 element was integrated upstream of the poly(A) tail to enhance the expression and translation of the entire cassette. Cloning of the plasmids was done individually and confirmed by Sanger sequencing or whole plasmid sequencing. The gRNA design was completed using the online platform https://www.benchling.com. The packaging of AAV viruses and titration were performed by PackGene Bioscience.
FACS
First, we perfused each mouse with 1× PBS and isolated brain cortex tissue. The tissue was treated using a neural tissue dissociation kit (Miltenyi Biotechnology) to fully lyse the cells and to isolate single cells. Then, we used a BD Fusion FACS flow cytometer (BD) to sort and collect the single cells. Note that for each sorting and collection experiment, we maintained consistent parameters, such as voltage. We also used the 488–513 nm fluorescence channel to sort GFP-positive cells and collected single cells with fluorescence intensity above log103.
Nested PCR
gDNA was extracted from tissue samples using a standard phenol–chloroform method, followed by quantification with a NanoDrop spectrophotometer. The initial PCR amplification was performed using outer primers designed to flank the target region, with a reaction mixture containing 50 ng gDNA, 0.5 μM each of outer primer, 10 μl of 2× Ex Taq polymerase mix and 7 μl nuclease-free water in a total volume of 20 μl. Thermal cycling conditions included an initial denaturation at 95 °C for 5 min, followed by 20 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 10 min. For the nested PCR, the first-round PCR product was diluted to 1/1,000 of its original concentration as the template, using inner primers specific to the target sequence. Cycling conditions were identical to the initial PCR. Amplified products were analysed by agarose gel electrophoresis stained with ethidium bromide, and bands were visualized under UV light for subsequent sequencing. Raw gel images of the PCR products are shown in Extended Data Fig. 6d and in Supplementary Fig. 1.
Analysis of on-target editing efficacy and bystander effect in cells
In brief, for the in vitro single-base editing efficiency and bystander effect analysis, we used the EditR online analysis tool (https://moriaritylab.shinyapps.io/editr_v10/) to analyse and quantify the Sanger sequencing chromatograms48. For the in vivo base-editing efficiency analysis, we first extracted gDNA from the single cells sorted by flow cytometry. Then, we constructed the NGS library using different index sequences. The library samples were sent to Shanghai JMDNA Biology. After obtaining the sequencing data, we used CRISPResso2 to analyse and quantify the results with support from the Medical Science Data Center of Fudan University.
GUIDE-seq
GUIDE-seq experiments were performed in human HEK293T and mouse Neuro-2a cells following established protocols49 with minor modifications. In brief, 3 × 105 HEK293T or Neuro-2a cells were electroporated with Cas9–sgRNA ribonucleoprotein (RNP) complexes—comprising 5 µg WT Streptococcus pyogenes Cas9 protein and 2 µg CHD3-targeting sgRNA (5′-TTGAGTCTCCACAGCATGTG-3′)—together with 1.5 µg annealed double-stranded oligodeoxynucleotide (dsODN). Electroporation was performed using a Lonza 4D-Nucleofector system (Program DG-130). After 48–72 h, cells were collected and gDNA was extracted using a TIANamp Genomic DNA kit (Tiangen) for GUIDE-seq analysis (Azenta Life Sciences) using a previously described computational pipeline49. Because the engineered CHD3(R1025W/R1025W) cell line displayed markedly reduced viability following electroporation, WT HEK293T cells were used for GUIDE-seq. The specific off-target sites identified by GUIDE-seq in HEK293T cells are listed in Supplementary Table 10. HEK293T and Neuro-2a cells were obtained from the Cell Bank of the Chinese Academy of Sciences. Regular short tandem repeat (STR) profiling for cell line authentication and mycoplasma testing were performed by the Cell Bank of the Chinese Academy of Sciences.
sgRNA sequences
Sequences of the sgRNAs C10, C11, A4, A10, A11, A13 and A15 (Extended Data Fig. 4) are provided in Supplementary Table 11.
R-loop assay
HEK293T cells were plated, cultured and transfected in a 24-well plate with 500 ng each of dSaCas9–sgRNA plasmid carrying a mCherry cassette and the tested ABE plasmid carrying a GFP cassette. After culturing for another 72 h, GFP–mCherry double-positive cells were sorted (BD FACSAriaFusion flow cytometer) and lysed using DirectPCR reagent (Viagene Biotech). Targeted PCR was performed using LA Taq (Takara) for high-throughput sequencing (Decode Biomedicine Technology).
Analysis of on-target editing efficacy, bystander effects and off-target effects in mice and monkeys
In mice, off-target sites were determined using GUIDE-seq. Primers used are provided in Supplementary Fig. 4 and Supplementary Table 12. The list of all identified genomic sites with read counts before and after filtering and consolidation are provided in Supplementary Table 13.
In monkeys, off-target sites were predicted using the Benchling CRISPR design tool, with the top ten predicted off-target sites listed in Supplementary Table 4. gDNA from brain tissues was extracted, and nested PCR was performed to amplify regions containing on-target sites and potential off-target sites. The primers used are detailed in Supplementary Table 14. Amplified products were submitted to Suzhou Hongxun Biotechnologies for NGS. Sequencing libraries were quantified and sequenced on an Illumina NextSeq 500/550 High Output Kit v.2.5 (75 cycles).
Immunohistochemistry
For immunofluorescence staining of cultured primary neurons, the cell medium was discarded and cells were washed with 1× PBS. Next, 4% paraformaldehyde (PFA) diluted with 1× PBS was used to fix cells for 20 min at room temperature. Following fixation, cells were washed with 1× PBS and blocked in a solution containing 5% BSA and 0.4% Triton X-100 in 1× PBS for 2 h at room temperature. Following fixation, cells were washed with 1× PBS and blocked in a solution containing 5% BSA and 0.4% Triton X-100 in 1× PBS for 2 h at room temperature. Data analyses of Extended Data Fig. 1m were collected and presented in Supplementary Table 15.
For immunofluorescence staining of Cas9 in mice, 0.8% Triton X-100 was used. Cells were incubated with primary antibodies prepared in the same blocking solution overnight at 4 °C, followed by secondary antibody incubation for 3 h at room temperature. For brain tissue preparation, mice were transcardially perfused with 1× PBS, followed by 4% PFA. Brains were post-fixed in 4% PFA overnight and dehydrated in 30% sucrose in 1× PBS. Dehydrated brains were sectioned into 40-μm slices using a Leica CM1950 cryostat. Sections were washed with 1× PBS, blocked with 5% BSA and 0.4% Triton X-100 in 1× PBS for 2 h at room temperature and incubated with primary antibodies overnight at 4 °C. Secondary antibody incubation was performed for 1 h at room temperature. Images were acquired using an Olympus VS200 high-throughput fluorescence microscope or an Olympus FV3000 confocal fluorescence microscope.
Nissl staining
Brains were collected from mice following euthanasia and fixed in 4% PFA overnight at 4 °C. After fixation, tissues were dehydrated through a graded ethanol series (70%, 95% and 100%) and embedded in paraffin. Coronal sections were cut at a thickness of 5 μm using a microtome and mounted onto glass slides. Deparaffinization was performed by immersing slides in xylene, followed by rehydration through descending ethanol concentrations and a final rinse in distilled water. Sections were then stained with 0.1% cresyl violet solution for 10 min at room temperature, followed by differentiation in 95% ethanol until the desired contrast was achieved. Slides were dehydrated again through ascending ethanol series, cleared in xylene and coverslipped with a mounting medium. Images were acquired using an Olympus VS200 high-throughput fluorescence microscope.
Antibodies
The primary antibodies used in this study and their dilutions are as follows: rabbit anti-CHD3 (1:1,000 dilution, ab109195, Abcam); rabbit anti-GAPDH (1:3,000 dilution, D264398, Sangon Biotech); rabbit anti-CRISPR–Cas9 (1:500 dilution, ab204448, Abcam); chicken anti-NeuN (1:1,000 dilution, ABN91, Millipore); rabbit anti-Flag (1:3,000 dilution, D110005, Sangon Biotech); rabbit anti-TBR1 (1:500 dilution, ab183032, Abcam); rat anti-CTIP2 (1:500 dilution, cab18465, Abcam); mouse anti-SATB2 (1:500 dilution, ab51502, Abcam); rat anti-CUX1 (1:500 dilution, ab307821, Abcam); goat anti-SOX2 (1:500 dilution, AF2018, Biotechne); rat anti-KI67 (1:500 dilution, 14-5698-82, ThermoFisher); goat anti-doublecortin (1:500 dilution, SC-8066, SANTA); rabbit anti-parvalbumin (1:500 dilution, ab181086, Abcam); donkey anti-DAPI (1:1,000 dilution; 28718-90-3, Sigma-Aldrich); donkey anti-rabbit IgG Alexa 488 (1:1,000 dilution; A32790; Thermo Fisher Scientific); donkey anti-rabbit IgG Alexa 555 (1:1,000 dilution; A32794; Thermo Fisher Scientific); donkey anti-mouse IgG Alexa 488 (1:1,000 dilution; A32766; Thermo Fisher Scientific); donkey anti-mouse IgG Alexa 555 (1:1,000 dilution; A32773; Thermo Fisher Scientific); donkey anti-rat IgG Alexa 555 (1:1,000 dilution; A48270; Thermo Fisher Scientific); donkey anti-goat IgG Alexa 555 (1:1,000 dilution; A32816; Thermo Fisher Scientific); and donkey anti-chicken Alexa Fluor 488 (1:1,000; 20166; Biotium).
RNA-seq
Brains were collected from mice and preserved in RNA later solution. The library samples were sent to Suzhou Hongxun Biotechnologies for RNA-seq. mRNA was transformed into cDNA using a TruSeq RNA Sample Prep kit v.2 (Illumina), and sequencing was carried out on an Illumina NovaSeq 6000 platform. The RNA-seq data were used to correct the full-length transcriptome sequence of mice through LoRDEC software. Differential expression analysis was performed using the DESeq2 R package (v.1.20.0)50. Gene ontology enrichment and KEGG analysis of differentially expressed genes was implemented by the clusterProfiler R package, in which gene length bias was corrected51.
ATAC–seq
Brain tissue was collected from mice at 10 weeks of age and immediately frozen in liquid nitrogen to preserve cellular integrity. The library samples were sent to Suzhou Azenta Life Science for ATAC–seq. Nuclei were isolated and subjected to a single PBS wash before lysis52. Nuclei were then directly processed with transposase to tagment accessible chromatin DNA, incorporating partial adapter sequences at the ends of fragmented DNA. Subsequent amplification enriched the library, with full Illumina sequencing adapters introduced to complete library construction. The resulting libraries were initially quantified and diluted using a Qubit fluorometer, followed by assessment of insert size and nucleic acid concentration with an Agilent 2100 Bioanalyzer. After pooling, the effective concentration of the combined libraries was precisely determined via qPCR to ensure accurate sequencing loading concentration and reliable data output. All computational analyses were conducted using a high-performance computing cluster, with results visualized using DESeq2 R package and IGV for downstream interpretation53.
Intravenous tail injection of AAVs in mice
To investigate the effect of ameliorating the aberrant phenotype in Chd3hR1025W/+ mice after delivery of the TeABE system, AAV-PHP.eB-hSyn-GFP, AAV-PHP.eB-hSyn-nCas9-1-1029 and AAV-PHP.eB-hSyn-nCas9-1030-1248-TadA-F148A-nCas9-1264-1368-U6-sgRNA were packaged by PackGene Biotech. Virus titers were quantified by qPCR, and infectivity was evaluated by measuring fluorescence intensity following infection. Anaesthetized 6-week-old Chd3hR1025W/+ mice (for the treatment group) were injected with a mixture of AAV-PHP.eB-hSyn-nCas9-1-1029 (100 μl of 5 × 1012 vg ml−1), AAV-PHP.eB-hSyn-nCas9-1030-1248-TadA-F148A-nCas9-1264-1368-U6-sgRNA (100 μl of 5 × 1012 vg ml−1) and AAV-PHP.eB-hSyn-GFP (10 μl of 5 × 1012 vg ml−1) into the tail vein. Chd3+/+ mice (for the blank control group) and Chd3hR1025W/+ mice (for the negative control group) were injected with AAV-PHP.eB-hSyn-nCas9-1-1029 (100 μl of 5 × 1012 vg ml−1), AAV-PHP.eB-hSyn-nCas9-1030-1248-TadA-F148A-nCas9-1264-1368-U6 (100 μl of 5 × 1012 vg ml−1) (no-targeting TeABE) and AAV-PHP.eB-hSyn-GFP (10 μl of 5 × 1012 vg ml−1).
Behavioural tests
The age of the mice subjected to behavioural tests and the P values for all behavioural experiments are detailed in Supplementary Table 6. These mice were handled for 4 days before testing. Behavioural data were recorded and analysed using Ethovision XT software (research resource identifier: SCR_000441, Noldus), with investigators blinded to the genotypes of the mice. Before testing, mice were habituated to the behavioural test room for at least 1 h to acclimate to the environment. The apparatus was thoroughly cleaned, and any residual odours were removed with deodorizer before and after each trial. During the tests, mice were allowed to move freely and explore the apparatus. All behavioural tasks were performed between 9:00 and 18:00, corresponding to the light phase of the light–dark cycle. Light intensity was standardized to 50 lux for the open-field, self-grooming, buried-marble and home-cage intruder social-interaction tests. An intensity of 80 lux was used for the three-chamber social interaction, novel object recognition, tail suspension, joint analysis, gait analysis, stride analysis and rota-rod test. The maximum light intensity was set to approximately 300 lux as an aversive stimulus for the elevated-plus maze and Barnes maze. Behavioural tests were conducted using protocols generally consistent with a previous report29.
USV test
USVs were elicited through a rapid maternal separation procedure conducted on pups at P3. To record USVs, pups were individually placed into a clean plastic container (containing bedding material) and subsequently placed inside a clean polystyrene container. The lid of the container was securely closed to minimize external noise and testing began immediately after placement. Each trial lasted for 5 min. The USVs were detected using an ultrasonic microphone (Ultravox Noldus), connected via the Ultravox device (Noldus) positioned near the pup’s location. Data were analysed using DeepSqueak (v.2.6.2), and the investigator conducting the data analysis was blinded to the genotypes of the mice.
Open-field test
The open-field test was performed in a custom-built apparatus made from 0.75-cm-thick white plastic, measuring 40 × 40 × 40 cm. At the start of each trial, a single mouse was placed at the centre of the box and allowed to explore freely for 10 min. Behaviour was recorded using a Da Hua high-definition video camera. For analysis, the total distance moved, average velocity, movement tracks and time spent in the centre zone (20 × 20 cm) were assessed using Ethovision XT software, and results were visualized with ImageJ. The investigator conducting the data analysis was blinded to the genotypes of the mice.
Buried marble test
A symmetrical 6 × 6 grid of 36 black glass marbles was arranged on 7 cm of bedding in a clean standard mouse cage measuring 40 × 40 cm2. Mice were introduced into the cage and allowed to explore for 10 min. Photographs were taken at the start and end of the exploration period to determine the number of marbles buried. A marble was considered buried if more than 50% of its surface was covered by bedding.
Social-intruder test
The task was conducted in the home cage of the experimental mouse, with the feed trough, food and water bottles removed. Mice were individually housed for 5 days to ensure social isolation and were acclimated to the experimental environment for 1 h before the task. The social recognition task consisted of two phases: a social approach and familiarity session, followed by a social novelty and recognition session. During the social approach and familiarity session, a stranger mouse (mouse 1; C57BL/6J adult male, 6 weeks old) was introduced into the cage for 3 min, allowing free exploration and interaction. This was followed by three additional trials with 5-min inter-trial intervals to facilitate familiarity with mouse 1. In the social novelty and recognition session, a new stranger mouse (a littermate of mouse 1) was introduced into the cage. Cumulative sniffing time was recorded to analyse social interaction, specifically when the test mouse actively sniffed or made physical contact with the stimulus mouse via nose touching. The recognition index was calculated as the difference in cumulative sniffing time between trials T5 and T4. Video recordings were captured using a Da Hua high-definition camera, and cumulative sniffing time was measured with a split-second chronograph. The investigator conducting data analysis was blinded to the genotypes of the mice.
The three-chamber social-interaction test
The three-chamber test was conducted using a behavioural apparatus made of 0.75-cm-thick white plastic (dimensions: 60 × 40 × 30 cm3). Each chamber was equipped with 4 × 4 cm2 cut-out doors in the partition walls to permit free movement between the chambers. Small iron cages were placed in each side chamber to house the social partner mice. The day before the testing, mice were habituated to the apparatus for 1 h with empty cages in both sides of the chambers. Social partner mice (C57BL/6J adult male, 6 weeks old) were placed in the iron cages for 10 min to minimize stress and anxiety. The test comprised habituation, social approach and social novelty. On the day of the test, the subject mouse was placed in the centre chamber and allowed to freely explore all three chambers during the habituation session. During the social approach session, while the test mouse remained in the centre chamber, a social partner mouse (mouse 1) was placed in the iron cage in one of the side chambers while the other iron cage was empty. The test mouse was then allowed to explore the entire apparatus for 10 min. In the social novelty session, the test mouse was returned to the centre chamber, and a new stranger mouse (mouse 2) was placed in the iron cage in the opposite side chamber. The test mouse was allowed to explore for another 10 min. The apparatus was thoroughly cleaned with a deodorizer between trials to eliminate any olfactory cues. Video recordings were captured using a Da Hua high-definition camera, and the time spent interacting with the social partner or the empty cage (20 × 10 cm2) and locomotion heatmaps were analysed using Ethovision XT software and plotted with ImageJ. The investigator analysing the data was unaware of the genotypes of the mice.
Novel object recognition test
The novel object recognition task was carried out using an apparatus made of 0.75-cm-thick white plastic, measuring 24 × 24 × 24 cm3. One day before the test, each mouse was placed individually into the apparatus for 1 h to habituate to the environment. During the habituation session, each test mouse was placed in the centre of the apparatus and allowed to freely explore for 10 min. On the training day, which took place 1 day after habituation, the test mouse was again placed in the centre of the apparatus and allowed to freely explore for 10 min. On the recognition session, held 1 day after the training session, the test mouse was placed in the centre of the apparatus and allowed to freely explore for 10 min. In the object recognition session, one red cube (object 1) was placed at the top corners of the apparatus. Then the test mouse was taken out and the box was deodorized with the deodorant. In the novel object recognition session, the test mouse was again placed in the centre of the apparatus and allowed to freely explore for 10 min. In this session, the red cube (object 1) and one green cylinder (novel object) were placed at the top corners of the apparatus. Video recordings were captured using a Da Hua high-definition camera during the task. The time spent interacting with each object (within a nose point range of ≤3 cm) and the locomotion heatmaps were analysed using Ethovision XT software and plotted using ImageJ. The investigator analysing the data was unaware of the genotypes of the mice.
Elevated-plus maze test
The elevated-plus maze test was performed using a specialized behavioural apparatus. The closed arms were made of 0.75-cm-thick white plastic boards, each measuring 30 × 5 × 15 cm, whereas the open arms measured 30 × 5 × 15 cm. The total height of the apparatus was 50 cm. Each test mouse was placed at the centre of the maze and allowed to freely explore for 5 min to fully navigate the maze. Video recordings were captured with a Da Hua high-definition camera. The time spent in the open and closed arms was recorded, and data analysis was conducted using Ethovision XT software and plotted with ImageJ. The investigator performing the analysis was unaware of the genotype of each mouse.
Barnes maze test
The Barnes maze consisted of a circular, thick, white plastic board with a diameter of 122 cm, featuring 40 evenly spaced holes, each 5 cm in diameter, around its perimeter. The maze stood 80 cm above the ground and was designed to rotate around its centre. An escape cage, made of black lightproof plastic, was positioned beneath the target hole for accessibility. Two bright supporting lamps placed at opposite ends provided maximum light intensity as an aversive stimulus. Between each trial, the maze and escape cage were thoroughly cleaned with deodorizer to eliminate residual odours, and the maze was randomly rotated to avoid olfactory cues. The Barnes maze test included five consecutive days of training followed by two test sessions on the sixth and thirteenth day. Before training, mice were habituated to the environment for at least 1 h. During each trial, a mouse was placed at the centre of the maze under a dark opaque plastic cover for approximately 10 s and then released to explore. Each mouse had up to 3 min to locate the escape hole; if unsuccessful, the mouse was guided to the target hole and allowed to familiarize itself with the location for 1 min inside the escape box, which was covered with an opaque plastic shield to reduce light exposure. Each mouse underwent two training trials daily (10 trials in total over 5 days) to memorize the target hole location and learn to navigate to it independently. On the test day, the escape box was removed, and each mouse was allowed to explore the maze freely for 180 s. Video recordings were captured using Da Hua Smart video recording software. The primary latency to reach the target or opposite zone was calculated within 180 s, and locomotion traces were analysed using Ethovision XT software and plotted with Image J. All analyses were performed by investigators who were unaware of the genotype of the mice.
Tail suspension test and joint laxity analysis
The tail suspension test was used to assess muscle tone in mice by observing limb clamping. Mice were suspended by their tails, and a high-definition digital camera was used to captured real-time images. The position of the limbs was monitored for 10 s. A scoring system was applied based on the limb posture: a score of 0 was assigned if both hindlimbs and forelimbs were consistently splayed outward, away from the abdomen; a score of 1 was given if one limb was retracted towards the abdomen for more than 50% of the suspension time; a score of 2 was assigned if half of the limbs were partially retracted towards the abdomen for more than 50% of the time; and a score of 3 was given if the three limbs were fully retracted and touching the abdomen for more than 50% of the time; and a score of 4 was given if both hindlimbs and forelimbs were fully retracted and touching the abdomen for more than 50% of the time54. The clasping scores of the mice were recorded and analysed with all analyses performed by investigators blinded to the genotypes of the mice.
For the analysis of joint laxity: a score of 0 indicated normal joints with no observable redness or swelling; a score of 1 indicated mild redness and swelling localized to the ankle or wrist, or limited redness and swelling confined to individual digits; a score of 2 indicated for moderate redness and swelling affecting the ankle or wrist; a score of 3 indicated severe redness and swelling involving the entire paw, including the digits; a score of 4 indicated maximal inflammation extending to multiple joints within the limb55. To ensure objectivity, all joint laxity scores were recorded and analysed by investigators blinded to the genotypes of the mice.
Gait analysis and stride analysis
Analysis was performed as in ref. 56. Step pattern tracking was performed using a tunnel constructed from three pre-cut clear acrylic panels, each 10 mm thick, allowing mice to walk comfortably and take at least four steps. The hindlimb foot angle and spacing were recorded using an electronic camera taken from their abdomen. Thick, smooth paper, such as watercolour paper, was cut into strips slightly larger than the tunnel dimensions (approximately 380 mm long and 90 mm wide). Two contrasting non-toxic, washable, water-based paints (for example, blue and red) were applied to distinguish between hindlimb (blue) and forelimb (red) steps. The selected paints were safe for ingestion, as mice tended to lick residual paint off their feet after testing. Step intervals were recorded for subsequent analysis.
Rotarod test
This test requires mice to balance on a rotating rod, and their latency to fall is recorded as the end-point measure57. The rotarod consists of a circular rod turning at a constant or increasing speed. Animals placed on the rotating rod try to remain on it rather than fall onto a platform some 30 cm below. Before the test, all mice are trained for 1 day to adapt to the rotarod instrument. On the test day, the mice are placed on the rotating lane of the rotarod instrument and the test starts. The time of the mice staying on the stick and the final velocity is recorded while the investigator conducting data analysis was blinded to the genotypes of the mice.
Intrathecal injections of AAV in monkeys
All experimental protocols and animal care and handling were approved by the Ethics Committee of the Kunming Medical University (Kmmu20205DS). All monkeys were pair-housed in accordance with standard operating procedures in a climate-controlled room at 18 °C to 26 °C with a relative humidity of 40–70%, a 12-h light–hour dark cycle, and a ventilation rate of eight times per hour. Monkeys had free access to drinking water and were continually fed with monkey chow (4% calories from fat, 16% calories from protein and 80% calories from carbohydrates) at 200–300 g per day. Monkeys were also provided a daily allotment of fruits, vegetables, or additional supplements and various toys.
Monkeys were anaesthetized with an intramuscular injection of ketamine hydrochloride (5 mg kg−1) followed by an intravenous injection of propofol (1 mg kg–1) to induce and maintain anaesthesia. Following anaesthesia induction, the lumbar area was shaved and disinfected with povidone–iodine. The animals were positioned in lateral recumbency on the surgical table with the lumbar spine flexed to facilitate access to the intervertebral spaces. A spinal needle was inserted at an approximately 70° angle to the dorsal midline axis into the intervertebral space between L3 and L4. The needle advanced through the skin and subcutaneous tissue into the subarachnoid space, which was identified by a distinct loss of resistance and confirmed by the appearance of cerebrospinal fluid after gentle aspiration. The mixture of dual AAV9 vectors (1,200 μl of 1.00 × 1014 vg per ml of each vector for high dose, 300 μl of 1.00 × 1014 vg per ml of each vector for low dose) was then administered slowly into the subarachnoid space over a period of 60 s58. Following the injection, the needle was removed, and the site was closed and disinfected. Monkeys were monitored post-procedure for recovery and potential adverse effects.
Immunohistochemistry staining for monkey samples
Monkeys were anesthetized with ketamine hydrochloride (veterinary drug approval number (2015)100761663 by the Ethics Committee of the Kunming Medical University; 10 mg kg–1, intramuscular) and pentobarbital (45 mg kg–1, intramuscular) and transcardially perfused with 2,000 ml of 4 °C PBS and 500 ml of 4% PFA (Sigma-Aldrich, 16005) in PBS. After perfusion, the hemispheres of the brain were dissected, cut into small blocks (segmentation of subregion for monkey brain were selected and collected by a skilled technician with over a decade of experience), fixed with 4% PFA in PBS. The fixed brain tissue blocks were then cut into 40-μm cortical sections with a Leica Biosystems (Leica CM1950). Sections were washed for 5 min in PBS containing 5% BSA and 2% Triton X-100, and incubated with primary antibodies (in PBS with 1% BSA and 2% Triton X-100) overnight at 4 °C and subsequently with corresponding secondary antibodies. DAPI (1:1,000 dilution; 28718-90-3, Sigma-Aldrich) was used to label the nuclei and sections were mounted with 75% glycerol. A classical rabbit anti-Cas9 antibody (1:500 dilution, ab204448, Abcam) was used to label Cas9.
Statistical analysis
All statistical data are presented as the mean ± s.d., with n representing the number of individual experiments or mice. Statistical analysis was conducted using Prism (v.6.01, GraphPad Software) and Origin (v.2019b) to assess group differences. The significance of differences between groups was evaluated using a paired or unpaired two-tailed Student’s t-test, one-way repeated-measures ANOVA or two-way ANOVA, as indicated in the figure legends. A paired two-tailed t-test was used to compare the cumulative time spent interacting with the mouse during the social approach and social novelty sessions in the three-chamber test for each mouse. A one-way repeated-measures ANOVA was used to analyse differences among more than two groups under a single-factor assumption. Two-way ANOVA was applied to examine interactions between two factors. An unpaired two-tailed t-test was used in other comparisons. Significance was determined as follows: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. Sample sizes were determined before the experiments without using any statistical methods. Mice were randomly assigned to experimental groups. Data collection and analyses were not conducted in a blinded manner except for the immunohistochemical and behavioural tests, which were performed blind to mouse genotype. Data distribution was assumed to be normal, although this was not formally tested.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.