Cell lines and primary cell cultures
All cells were cultured under 5% CO2 at 37 °C. HEK-293T and COS7 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 1% l-glutamine and 1% penicillin–streptomycin (Sigma). HUDEP-2 cells (provided by Y. Nakamura and R. Kurita) were cultured as described21.
CD34+ haematopoietic stem and progenitor cells were isolated from peripheral blood from donors and sorted using CD34+ MACS (Miltenyi) according to the manufacturer’s protocol. The cells were differentiated using a three-phase erythroid in vitro differentiation protocol as previously described52,53. In brief, the cells were expanded in StemSpan medium (Stem Cell Technologies) supplemented with 100 ng ml−1 stem cell factor (SCF) (PeproTech), 100 ng ml−1 Flt-3 (PeproTech), 100 ng ml−1 thrombopoietin (TPO) (PeproTech) and 20 ng ml−1 IL-3 (PeproTech) for a total of six days.
After six days, the cells were reseeded in phase I differentiation medium (days 1–7) consisting of Iscove’s modified Dulbecco’s medium (IMDM) (Gibco) supplemented with 2% human plasma (Stem Cell Technologies), 3% human serum (Sigma), 3 units ml−1 heparin (Sigma), 10 µg ml−1 insulin (Sigma), 3 units ml−1 erythropoietin (Janssen-Cilag), 10 ng ml−1 SCF (PeproTech), 1 ng ml−1 IL-3 (PeproTech) and 200 µg ml−1 holo-transferrin (Sigma), at a density of around 0.5 × 106–1 × 106 cells per ml for a total of seven days. Phase II medium (days 8–12) included the same cytokines except for IL-3. During phase III (days 13 and beyond) holo-transferrin was increased to 1 mg ml−1 and SCF was withdrawn. Erythroid differentiation was monitored by fluorescence-activated cell sorting (FACS) analysis and May–Grünwald–Giemsa (RAL Diagnostics) staining of cytospin slides. Images were captured from cytospin slides using a PrimoStar 3 microscope (Zeiss) coupled with an Axiocam 208 camera (Zeiss). For growth-curve analysis, cultured erythrocyte progenitor cells were seeded in the same amounts and then counted at the indicated days both by haemocytometer (Tali, Invitrogen) and by microscope with trypan blue staining (Invitrogen) to exclude dead cells. To test for TGFβ stimulation, HUDEP-2 cells were stimulated with 5 ng ml−1 TGFβ (Sigma) and sampled for SMAD3 and CCND3 mRNA expression profiling at various time points after the treatment (0 min, 60 min, 120 min, 240 min and 300 min). The inhibitory effects on this pathway were evaluated with 20 μM SB-505124 (Sigma) in 0.2% DMSO, as previously described54. All of the cells used tested negative for mycoplasma contamination.
Human blood samples
Whole-blood samples were collected from volunteers enrolled in the SardiNIA general population cohort14, at the project centre in Lanusei (Ogliastra, Sardinia). All participants gave informed consent for the study protocols, which were approved by the Sardinian Regional Ethics Committee (protocol 2171/CE). These participants have agreed in principle to be recalled for functional follow-up experiments based on their genotypes at loci of interest, such as those reported in this study. The samples collected for the experiments reported in this study were specifically selected to ensure representation of homozygous genotypes at the loci of interest, rs112233623 and rs9349205 close to the CCND3 gene. For the malaria experiments, individuals who were carriers of the most frequent allelic variants in Sardinia that have previously been associated with protection against malaria were excluded from the collection. These variants include the β039 thalassaemia mutation (rs11549407-A) in the HBB gene (frequency = 0.051), the 3.7-kb α-thalassaemia deletion (NG_000006.1:g.34164_37967del3804; frequency = 0.14) and the Mediterranean deficiency variant (rs5030868-A) in the G6PD gene (frequency = 0.083), with the exception of a small group of samples that were selected to be hemizygous for the G6PD Mediterranean deficiency variant. We also tried to ensure a balance in terms of sex in the various genotype groups being compared (Supplementary Information).
Whole blood was collected using heparin vacutainer tubes (BD Biosciences) for gene-expression and cell-cycle experiments based on the generation of erythrocyte progenitor cells, and acid–citrate–dextrose (ACD) vacutainer tubes (BD Biosciences) for P. falciparum infection experiments. The blood was kept at ambient temperature in vitro before processing for the specific experiments, all of which began on the same day. Further details about sample collection are provided in the Supplementary Information, and the overlap of donors across the functional and P. falciparum growth experiments is detailed in Supplementary Table 5.
Flow-cytometry staining
All staining steps were performed at 4 °C, with efforts made to minimize sample exposure to ambient light. For flow-cytometry analysis, 3 × 105 in-vitro-differentiated erythroid progenitor cells were washed twice in PBS and resuspended in staining buffer (1% FBS in PBS). Cells were surface-stained for 20 min with the following antibodies: 1:50 PC7-conjugated CD235a (glycophorin A, Beckman Coulter), 1:100 PE-conjugated anti-Band3 (IBGRL) and 1:20 APC-conjugated anti-CD49d (integrin α4 chain, Beckman Coulter). After washing, cells were resuspended in 500 μl FACSFlow (BD Biosciences). For the single-cell population, CD235a–PC7 versus FSC-A was used to gate on CD235a–PC7-positive cells. Erythroblast maturation was evaluated by flow cytometry through the simultaneous detection of surface markers integrin α4, glycophorin A (GPA) and Band 3, using anti-CD49d–APC, anti-CD235a–PC7 and anti-Band3–PE monoclonal antibodies, respectively. Data were collected using BD FACS Accuri C6 Plus (BD Biosciences). For cell-cycle analysis at each time point, cultured erythroid progenitor cells were collected, washed in PBS, resuspended in permeabilizing solution and incubated at 37 °C for 30 min. Propidium iodide (BD Biosciences) was then added to a final concentration of 10 μg ml−1, and cells were incubated for an additional 10 min at room temperature before analysis. Cells were acquired on a FACSCanto (BD Biosciences) coupled with FACSDiva software (v.6.1.3).
The data analysis was performed using FlowJo software (v.10.10.0).
Luciferase reporter analysis
Firefly luciferase reporter constructs (pGL4.23, Promega) were generated by cloning the variant of interest centred in 372 nucleotides of genomic context upstream of the minimal promoter. Expression plasmids were prepared by subcloning human cDNA clones for each of SMAD3, p300 (clone provided by D. Barilà) and GATA1 (clone provided by F. Grosveld) into the pEF5-HA vector (vector provided by N. Bottini). The expression plasmid pCMV6-Entry FOG1 was purchased from Origene. To avoid erroneous normalization of transfection efficiency, we used a Renilla mutant control plasmid (pRLSV40-mut-GATA) in which the GATA response elements had been mutagenized, thus rendering pRL-SV40 unresponsive to GATA stimulation. Primer sequences are listed in Supplementary Table 4.
Firefly constructs (2,000 ng) were cotransfected with the pRLSV40-mut-GATA construct (300 ng) alone or with the expression plasmid (1,000 ng) into 625,000 HUDEP-2 cells using the Neon electroporation system (Invitrogen, 1200 V, 20 msec, two pulses). After 48 h, luciferase activity was measured using a Synergy 2 plate reader (Biotek) with a Dual-Glo luciferase assay system (Promega) according to the manufacturer’s instructions. Firefly luciferase levels were normalized to the pRLSV40-mut-GATA Renilla luciferase activity for each sample, and all values were then plotted relative to the empty pGL4 vector.
EMSAs
EMSAs were performed as previously described55. Radiolabelled double-stranded oligonucleotides were prepared by annealing oligonucleotides containing the rs112233623 WT (C) or the derived (T) allelic variants. Small-scale nuclear extracts were prepared as previously described56 from COS7 cells that had been treated with TGFβ (5 ng ml−1 for 16 h) as previously described57 and then transfected with the eukaryotic expression vectors pEF5HA-SMAD3 or pEF5HA-GATA1, or empty pEF5HA vector as a control. The presence and correct size of the proteins of interest in all extracts were verified in western blot assays. For supershift experiments, EMSA reactions were preincubated for 30 min on ice with specific antibody (anti-HA.1, BioLegend; anti-GATA1, Santa Cruz Biotechnology). Competitor cold oligonucleotides were added in 25×, 50× and 100× molar excess. The protein–DNA interaction products were resolved on a 5% non-denaturing acrylamide gel (37.5:1 acrylamide:bis-acrylamide, 0.25× TBE). Representative full-scan blots are provided in the Supplementary Information and in the source data. Primer sequences are listed in Supplementary Table 4.
ChIP assay
ChIP was performed by a SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads, Cell Signaling Technology, 9005S). In brief, cells (4 × 106 per immunoprecipitation) were fixed with 1% formaldehyde for 10 min at room temperature, and the reaction was quenched with glycine at a final concentration of 125 mM. Nuclei were prepared, and chromatin was incubated with micrococcal nuclease at 37 °C for 15 min, which was followed by an appropriate amount of sonication. Immunoprecipitations were performed using 10 μg of an antibody against SMAD3 or GATA1, or 1 μg with nonspecific rabbit IgG as negative control (2729S Cell Signaling Technology), according to the manufacturer’s instructions, at 4 °C for 12–16 h. The next day, the immunocomplexes were rotationally incubated with 30 µl of ChIP-Grade Protein G Magnetic Beads for two hours at 4 °C and then washed three times using low-salt wash buffer and once with high-salt wash buffer at 4 °C for 5 min per wash. Chromatin was eluted by ChIP elution buffer for 30 min at 65 °C with gentle vortex mixing (1,200 rpm) and cross-links were reversed by treatment with 5 M NaCl and proteinase K for two hours at 65 °C. ChIP DNA was purified and subsequently quantified by qPCR using primers designed to amplify a region surrounding rs112233623 in the CCND3 enhancer and SimpleChIP Human α Satellite Repeat Primers (4486, Cell Signaling Technology). Data were finally presented as percentages of the input DNA. Primer sequences are listed in Supplementary Table 4.
Genome editing by CRISPR–Cas9
Single guide RNAs (sgRNAs) targeting SMAD3 and SMAD2 were developed using a CRISPR design tool kindly provided by the Zhang laboratory (Broad Institute, MIT) (see Supplementary Table 4 for sgRNA sequences). The annealed oligos were ligated into the BsmbI-digested LentiViral (LV) construct LentiCRISPRV2 (Addgene). Lentiviral vectors were obtained by transfecting HEK-293T cells with 10 μg LentiCRISPRV2 (expressing both sgRNA and Cas9 or Cas9 only), 7.5 μg psPAX2, 2.5 μg pMD2G-VSVG and 1.25 μM pRSV-REV, using polyethylenimine (PEI; ref. 58) (Polysciences). After 72 h, viral particles in the supernatants were collected by centrifugation at 20,000 rpm for two hours at 4 °C. HUDEP-2 cells were then transduced with CRISPR–Cas9 by centrifugation at 2,800 rpm for 90 min at 30 °C with virus at a multiplicity of infection of 40. Forty-eight hours after transduction, cells were positively selected with the addition of 1 μg ml−1 of puromycin to the medium. To validate biallelic CRISPR indel detection at the genomic target site, DNA was extracted from puromycin-selected HUDEP-2 cells with the QIAamp DNA Mini Kit (QIAGEN), PCR-amplified (primer sequences are listed in Supplementary Table 4) and sequenced by the Sanger method in a 3130xl Genetic Analyzer (Applied Biosystems). The percentage of indels was calculated using TIDE v.3.3.0 (https://tide.nki.nl/)59.
RT–qPCR analysis
RNA was extracted from cells using the RNeasy extraction kit (QIAGEN) according to the manufacturer’s instructions. First-strand cDNA synthesis was performed with the SuperScript III kit (Thermo Fisher Scientific) for equivalent amounts of starting RNA from all samples. The cDNA was analysed on an ABI 7700 machine using SYBR green master mix (Applied Biosystems), and normalized to GAPDH or B2M (β2-microglobulin) as an internal control. All samples were done in triplicate. PCR cycle conditions were 95 °C for 5 min, and 40 cycles of 95 °C for 10 s, 54 °C for 10 s, 72 °C for 15 s. Analyses of Ct values were performed as previously described60. PCR primer pairs are listed in Supplementary Table 4.
Analysis of RNA-seq data from public datasets
Raw RNA-seq data (FASTQ files) of An et al.19 and Ludwig et al.20 were downloaded from the NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra). RNA-seq reads were aligned using STAR software (v.2.7.10b)61 against a transcriptome reference generated by RSEM software (v.1.3.1)62. The transcriptome reference was built from the hs37d5 human genome assembly (https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/) and GENCODE v.14 gene annotation (https://www.gencodegenes.org) using the rsem-prepare-reference function of RSEM. Normalized gene-expression levels (fragments per kilobase of transcript per million mapped reads; FPKM) were computed with the rsem-calculate-expression function of RSEM. Box plots were generated with R (v.3.5.3).
Western blotting
Western blotting was done following standard protocols. Proteins were collected in RIPA buffer (Millipore) supplemented with 2 mM PMSF (Sigma), and cOmplete Mini Protease Inhibitor Cocktail (Roche). Protein yield was determined with the Bradford protein assay (Bio-Rad), and equal amounts of total protein were separated by SDS–PAGE (Thermo Fisher Scientific). Proteins were transferred onto a polyvinylidene difluoride (PVDF; Amersham Biosciences) membrane in a wet transfer system (Bio-Rad). Membranes were then incubated with HRP-conjugated secondary antibodies (Santa Cruz Biotechnology), developed with an ECL system (Amersham Biosciences) according to the manufacturer’s instructions and visualized using photographic film. The ratio between cyclin D3 and β-actin was calculated using Image J1.52a (https://imagej.net/ij/). Representative full-scan blots are provided in the Supplementary Information and in the source data.
Reagents and antibodies
Recombinant TGFβ and TGFβ type I receptor inhibitor SB431542 were purchased from Sigma-Aldrich (T7039 and S4696). The following antibodies were used: PC7-conjugated anti-human CD235a (GPA–PC7, 1:50) supplied by Beckman Coulter, clone 11E4B-7-6, A71564; PE-conjugated anti-Band3 (Band 3–PE, 1:100) supplied by IBGRL, clone BRIC 6, 9439PE; APC-conjugated anti-CD49d (integrin α4–APC, 1:20) supplied by Beckman Coulter, clone HP2/1, B01682; anti-human GATA1 (GATA1, 1:10) supplied by Santa Cruz Biotechnology, clone N1, sc-266X; anti-influenza haemagglutinin epitope tag HA.11 (SMAD3–HA, 1:10) supplied by BioLegend, clone 16B12, MMS-101P; anti-human cyclin D3 (cyclin D3, 1:250) supplied by Santa Cruz Biotechnology, clone D-7, sc-6283; anti-human SMAD2 (SMAD2, 1:500) supplied by Santa Cruz Biotechnology, clone S-20, sc-6200; anti-β-actin (β-actin, 1:1,000) supplied by Santa Cruz Biotechnology, clone C4, sc-47778; anti-cyclin D3 (cyclin D3, 1:2,000) supplied by Cell Signaling Technology, clone DCS22, 2936S; rabbit anti-mouse IgG–HRP (1:20,000) supplied by Santa Cruz Biotechnology, sc2005; goat anti-rabbit IgG–HRP (1:20,000) supplied by Santa Cruz Biotechnology, sc2004; rabbit anti-goat IgG–HRP (1:20,000) supplied by Santa Cruz Biotechnology, sc-2768; anti-human SMAD3 (SMAD3, 10 µg in 100 µl for immunoprecipitation and 1:1,000 for western blot) supplied by Cell Signaling Technology, clone C67H9, 9523; anti-human GATA1 (GATA1, 10 µg in 100 µl) supplied by Abcam, ab11852; and normal rabbit IgG, (IgG, 1 µg in 100 µl) supplied by Cell Signaling Technology, 2729S.
Plasmodium falciparum culture
In vitro studies were done using the genetically stable reference FUP strain (mycoplasma-free) according to previously reported protocols63. The FUP strain of P. falciparum selected for our in vitro study (provided by E. Schwarzer) has been used extensively in both in vitro and in vivo studies to investigate various aspects of malaria infection, including RBC invasion, growth and development in the host. Compared with other strains, the FUP strain tends to maintain its genetic integrity during in vitro culture, making it a reliable choice for long-term studies64. To evaluate the reproducibility of our findings across genetically distinct parasite backgrounds, we performed additional experiments using the 3D7 (African) and DD2 (Southeast Asian) isolates.
Infection of RBCs from genotyped individuals
Fresh genotyped venous blood (about 25 ml) was collected from genotyped donors into ACD vacutainer tubes (BD Biosciences) and immediately processed. RBCs were separated from plasma and leukocytes by three washings in RPMI 1640 containing 25 mM HEPES (Gibco) and supplemented with 2 mM glutamine, 20 mM glucose, 27 μg ml−1 hypoxanthine and 32 μg ml−1 gentamicin (Sigma) (pH 7.2). The washed genotyped RBCs were used to maintain the parasite in vitro cultures in the same medium. Parasite cultures were synchronized as described previously65. Cultures were incubated at 37 °C under a 5% O2, 5% CO2, 90% N2 (v/v) atmosphere as previously described66.
To synchronize the genotyped cultures by parasite stage, schizonts (mature forms of Plasmodium) were collected from the 40–60% interface after passing a mixed-stage culture through a discontinuous Percoll mannitol density gradient by centrifugation at 5,000g for 30 min. The ring-stage parasites (early parasite stage) from the bottom fraction were also kept for further synchronous culturing. To start plasmodium cultures, density-isolated schizonts were added to erythrocytes in growth medium to a density of 2.5% parasites per erythrocyte.
All experiments involving in vitro malaria infections were performed in parallel with two initial haematocrit settings: 2% and 1%. This parallel experimental design provided the option to select between the two settings for subsequent short- and long-term parasite culture analysis, circumventing the complex and unpredictable process of repeating experiments from a small pool of donors with the relevant multilocus genotype combination, who are difficult to resample.
Samples with an initial haematocrit of 2% used 240 µl of RBCs with 2.5% schizont parasitaemia. We added 240 µl of packed erythrocytes every 48 h, coupled with replacement of the growth medium. This led to a gradual increase in haematocrit up to approximately 6% by the third cycle.
For 1% initial haematocrit samples, we used 2.5% schizont parasitaemia to infect 120 µl of packed erythrocytes and the same amount of RBCs was added to the cultures every 48 h, alongside the replacement of the growth medium. In this case, the final haematocrit level reached approximately 3% by the third cycle.
These two settings were used for all replicates at each time point (24, 48, 72, 96, 120 and 144 hpi). Because analysis of long-term cultures was performed to allow a wider range of observations, experiments were selected that began with an initial haematocrit level of 1%.
This is because the haematocrit level affects the glucose level significantly: as the haematocrit increases, so does glucose consumption, resulting in glucose depletion; in turn, the glucose level affects parasitaemia significantly, because the parasites rely heavily on glycolysis for energy and survival. Especially in the late cycles of long-term cultures, when the haematocrit level increases proportionally with the progressive addition of erythrocytes, we expect that parasitaemia will be less (negatively) affected by glucose consumption by both uninfected and infected erythrocytes at the selected lower haematocrit setting.
For each condition, two thin blood smears were independently counted by three trained microscopists in a blinded manner according to the latest guidelines of the World Health Organization (Malaria Microscopy Quality Assurance Manual). The resulting six counts were averaged to obtain a single arithmetic mean per condition.
Total parasitaemia, stages of morphology, parasite viability and inhibition rate were determined using Diff-Quick (RAL Diagnostics)-stained thin blood smears and light microscopy (Carl Zeiss Microscope Primo Star, objective lenses: 20×/0.50 PlanF and 100×/1.25 Oil Plan-ACHROMAT). The percentage of total parasitaemia was determined as follows: (number of parasitized erythrocytes/total number of erythrocytes) × 100. For each sample, at least 5,000 cells were counted. Damaged and pyknotic forms were counted separately and excluded from the total parasitaemia count.
The percentage of ring parasitaemia was determined as follows: (number of rings (single and multiple)/total number of infected RBCs) × 100.
The percentage of trophozoite parasitaemia was determined as follows: (number of trophozoites (single and multiple)/total number of infected RBCs) × 100.
The percentage of schizont parasitaemia was determined as follows: (number of schizonts (most mature form of P. falciparum asexual stages)/total number of infected RBCs) × 100.
The percentage of dead parasites was determined as follows: (number of pyknotic forms (blue dots inside RBCs)/total number of infected RBCs) × 100.
The percentage of parasite growth inhibition was calculated as follows: (1 – parasitaemia value)/(parasitaemia WT/WT|WT/WT average value) × 100.
The experiments were done in five batches to ensure, as far as possible, that individuals recruited for each experimental batch had a similar proportion of the different genotypes, especially IoE and DoE, to control for potential batch effects (Supplementary Information). A table listing the batch assignment for each donor used in the infection experiments is provided in the source data. We also ensured that all assays were performed under strictly controlled, standardized and homogeneous conditions to maintain consistency and comparability between batches, using exactly the same experimental protocols, including identical reagent batches, instrument settings and environmental parameters. This approach ensured the validity, accuracy and reproducibility of the results and minimized the variability associated with sample processing over time.
LC–MS measurements for ROS detection
Plasma was separated from whole blood by centrifugation at 200g for 5 min at 4 °C. White blood cells and pellets were thereby removed and the collected RBCs were transferred into a fresh microcentrifuge tube. The RBCs were then diluted 100-fold in PBS and probed with 20 μM coumarin boronic acid (Cayman Chemical). Before LC–MS analysis, 100 μl of methanol was added to the samples, which were then centrifuged at 5,000g for 30 min at 4 °C. LC–MS analysis was performed on an UltiMate 3000 UPLC system (Thermo Fisher Scientific) as previously described67. The limit of quantitation was calculated from calibration curves. For hydrogen peroxide species, the limit of detection was set to 11 nM, and the limit of quantification was set to 37 nM.
Statistical analyses
Targeted genetic association
Association signals for HbA2 and HbF levels (g dl—1), as well as MCV, RBC and MCH, were detected for the CCND3 gene region in a cohort of up to 6,824 samples from the SardiNIA cohort, as previously reported1 in a subset of 6,305 individuals from the same cohort in an analysis of haemoglobin levels.
Analyses used a genetic map based on 7,928 genotyped samples, with the OmniExpress Illumina array and iScan system using the Infinium HTS protocol, and imputed on a genome-wide scale using a Sardinian sequence-based reference panel of 3,514 individuals, as previously described51. The target region of ±0.5 Mb around the gene was analysed using a linear mixed model that accounts for cryptic relatedness and population stratification, implemented in the q.emmax function of the EPACTS program (http://genome.sph.umich.edu/wiki/EPACTS). Before the analysis, phenotype levels were normalized with inverse-normal transformation and adjusted for covariates of sex, age and age2 and for the presence of at least one of the 3 β0 mutations (β039 (rs11549407), HBB:c.20delA (rs63749819) and HBB:c.315+1G>A (rs33945777)), all directly genotyped or imputed. Moreover, HbF outliers with values higher than 5% were not considered in the association analysis, as previously reported1. An association signal was considered significant at a P-value threshold of 6.9 × 10−9 (ref. 51). Conditional analyses were performed for each trait by adding the most associated SNP as a covariate to the model adjusted for age, age2, sex and β0: no independent variant was found at the locus. To detect a causal variant, or causal variants, in the associated region, fine mapping was done by applying credible set analyses to the traits showing a significant association signal in the region (HbA2, MCV, MCH and RBC) using FINEMAP v.1.4.2 software; the –cond option and settings for one causal variant (–n-causal-snps 1) and for a maximum of ten causal variants (–n-causal-snps 10) for each association profile were used, as previously described68. Fine-mapping analyses converged for all tested traits after one causal SNP was detected, finding a single credible set at the 95% summed posterior probability. Variants in the credible set were then annotated for likely deleterious effect using the CADD score69 (using Ensembl GRCh37 release 111, January 2024).
Population genetic analyses
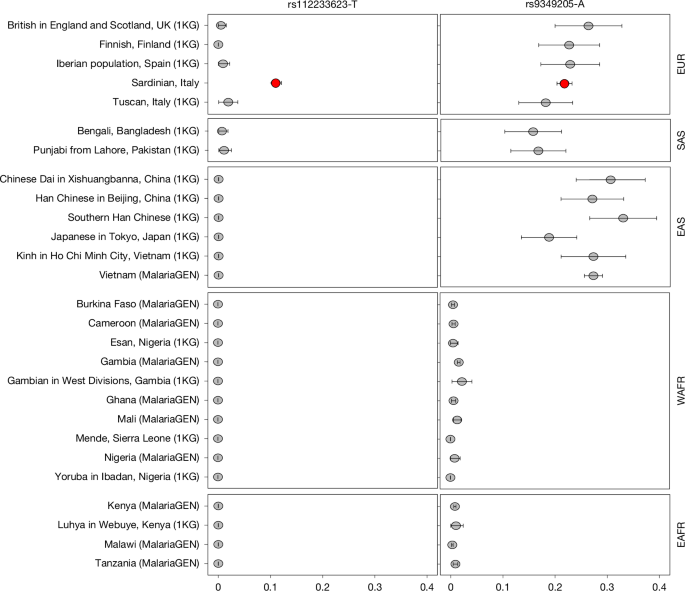
To investigate the hypothesis that the high allele frequency of rs112233623 in the Sardinian population results from positive selection rather than from genetic drift, standard metrics based on allele frequency (FST) and haplotype differentiation patterns (iHS and xp-EHH) were calculated as previously described70. In particular, a subset of 1,577 unrelated samples belonging to the SardiNIA whole-genome sequence reference panel51 was studied. This subset was defined by computing the genome-wide proportion of pairwise identity by descent (IBD) (π) on a random set of one million SNPs with a MAF > 0.05 in the 1KG population sample; for each pair of individuals with π > 0.05, the offspring was preferentially removed if in a trio; otherwise, the individual with the largest summed value of π across all other relationships with π > 0.05 was removed. Analyses on SardiNIA were then compared with those on 500 EUR samples included in 1KG phase 3 (ref. 18). To estimate the FST statistic, the Weir–Cockerham formula implemented in VCFtools v.0.1.12b (https://vcftools.github.io/index.html) was used to compare SardiNIA with 1KG EUR samples. For the haplotype-based analyses, variants with a MAF < 0.01 and a Hardy–Weinberg equilibrium (HWE) test P < 10−4 in Sardinian samples were first removed; this conservative filter facilitated better reconstruction of haplotypes by avoiding genotyping errors. The extended haplotype homozygosity (EHH) and the unstandardized iHS and xp-EHH values were then calculated using selscan71. Standardized statistics were obtained using the norm tool included in selscan. Finally, to establish a statistical benchmark for each test statistic, we constructed an empirical background distribution based on randomly selected variants from sequenced SardiNIA genomes that matched our target variant rs112233623 for three features: MAF ± 2% in SardiNIA; a measure of background selection (B score (ref. 72) ± 50 units); and local recombination rate in a 50-kb region around the variant (±0.5 cM per Mb; ref. 70) for a total of 13,134 variants. To exclude background variants in LD, variants with a correlation (r2) ≥ 0.1 were then filtered using the pruning procedure implemented in plink (https://www.cog-genomics.org/plink/1.9/), to obtain 1,075 variants that were used for the empirical background distribution used to calculate the genomic percentile values. Percentiles are calculated for standardized values for FST and xp-EHH tests, and for absolute standardized values for iHS. A high percentile value (such as 99%) suggests that for the statistic calculated, the variant shows more extreme signatures of positive selection than do 99% of the genomic background variants.
Coalescent-based analyses of recent positive selection
Description of dataset and preprocessing steps
For the purposes of coalescent-based analyses, we used the whole-genome sequencing dataset described previously73,74 (n = 3,514). This dataset includes individuals from the SardiNIA Project51, as well as from two case–control studies conducted in Sardinia. Related individuals were pruned from this dataset on the basis of the proportion of pairwise shared IBD, as described73. Given the computational demands, in a first step we sampled 100 unrelated individuals at random, followed by a second step in which we randomly selected a larger sample of 300 unrelated individuals for coalescent-based analyses.
Construction of genealogies using Relate
Genealogies were constructed using the Relate method (v.1.1.9), as described previously75. In brief, this method approximates the ancestral recombination graph by inferring a series of marginal trees along the genome. For all analyses using Relate, we followed the pipeline described in the online documentation (https://myersgroup.github.io/relate/). First, the ConvertFromVcf module was used to convert files from VCF format to the haps/sample format used by Relate, and the provided script (PrepareInputFiles.sh) was used to prepare the data. Then Relate was run with the mutation rate set to 1.25 × 10−8 and the haploid effective population size set to 30,000 to generate anc/mut files. The script EstimatePopulationSize.sh was then used to estimate coalescence rates, which were then used to re-estimate branch lengths in the anc/mut files with the script ReEstimateBranchLengths.sh. Marginal trees were visualized using the provided script TreeViewMutation.sh.
Genealogies for the sampled Sardinian individuals
We first constructed a genealogy for the 100 sampled individuals described in the ‘Description of dataset and preprocessing steps’ subsection above, using data from chromosome 6. As a precursor to using CLUES to infer selection coefficients across the genome, we repeated the Relate analysis using data from each autosome (1–22). We then used the script SampleBranchLengths.sh to sample branch lengths using Markov chain Monte Carlo (MCMC) for each variant in the SNP set of the 1,075 selected matching variants. We used the coalescence rates inferred by Relate and these sampled branch lengths as input and set the mutation rate to 1.25 × 10−8 per bp per generation. This step was repeated for each variant for which we estimated a selection coefficient (see below).
Multi-population genealogy
We also constructed a genealogy using Relate for a merged dataset including the 100 individuals described in the ‘Description of dataset and preprocessing steps’ subsection above, 107 Tuscan individuals from Italy18 and 108 Yoruba individuals from Ibadan, Nigeria18. We followed the same procedure described in the ‘Construction of genealogies using Relate’ subsection above to construct and visualize this genealogy for variants on chromosome 6, including rs112233623 and rs9349205.
Estimation of allele frequency trajectory and selection coefficient for rs112233623 using CLUES
We used the CLUES method31 to jointly infer the allele frequency trajectory and selection coefficient for the DoE variant (rs112233623). To do so, we followed the steps described above to run Relate tree inference for the sampled individuals described in the ‘Description of dataset and preprocessing steps’ subsection above and sample branch lengths from the trees. We then used the CLUES script inference.py to run CLUES with the sampled branch lengths (–times) and coalescence rates (—coal) outputted by Relate as input to the script. The CLUES-provided script plot_traj.py was then used to visualize the allele frequency trajectory.
Empirical distribution of selection coefficient estimates
To contextualize the results of the selection inference procedure, we repeated the CLUES analysis for the 1,075 SNPs selected to match by MAF, B score (background selection) and recombination rate with that of the DoE variant rs112233623. The HLA region was excluded from this SNP set. Of the 1,075 matched SNPs, 1,035 and 1,054 mapped onto the marginal tree inferred by Relate using 100 and 300 samples, respectively, and the remaining variants were excluded from further analysis.
Statistical analyses in experiments
On the basis of data distributions and sample sizes, parametric t-tests or non-parametric Wilcoxon–Mann–Whitney tests were applied to compare means or medians, respectively, and to establish statistical significance. To test a specific hypothesis, the one-tailed alternative-hypothesis test was used. Statistical analyses were performed using R v.3.5.3 (2019-03-11).
Correlations were calculated using the Pearson’s product moment correlation coefficient and P values were computed under the two-side hypothesis (cor.test function in R).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.