Protein expression and purification
Point mutations were made in WT VPI10463 TcdA and WT TcdB2 as described previously18,19,20. Primer information can be found in Supplementary Table 2. TcdA, TcdB and CspC were recombinantly expressed and purified as previously described18,41. Plasmid information for all antigens is provided in Supplementary Table 2. Plasmids encoding NTAs were codon-optimized versions of the candidate NTAs from the C. difficile R20291 background which were synthesized at Genscript into a pET47b(+) vector and included a C-terminal 6×-histidine tag for protein purification. C40 peptidase 2, FlgG, FlgE, FlgK, CspC and polysaccharide deacetylase were transformed into E. coli BL21 (DE3) STARs (Supplementary Table 2). To express each NTA, 12 l of lysogeny broth medium supplemented with 50 mg l−1 kanamycin were inoculated with an overnight culture to an optical density at 600 nm (OD600) of 0.1. Cells were grown at 37 °C and 220 rpm. Expression was induced with 1 mM Isopropyl-β-D-1-thiogalactopyranoside (IPTG) once cells reached an OD600 of 0.4–0.6. After 4 h, cells were centrifuged and the pellets were resuspended in 20 mM Tris (pH 8.0), 500 mM NaCl, 2% lysis mix (phenylmethylsulfonyl fluoride (0.1 mM), leupeptin (2 mg ml−1), pepstatin (2 mg ml−1), 2% DNase (2 mg ml−1) and 2% lysozyme (10 mg ml−1)). Bacterial suspensions were lysed three times using an EmulsiFlex C3 microfluidizer (Avestin) at 15,000 lb in−2. Lysates were centrifuged at 40,000g for 45 min at 4 °C. NTAs were initially isolated from supernatant using a Ni2+-affinity column (HisTrap FastFlow Crude; GE Healthcare). NTA eluents were further purified using an S-200 size-exchange column (GE Healthcare) in 20 mM HEPES (pH 6.9) with 50 mM NaCl on the ÄKTA Pure fast protein liquid chromatography system (Cytiva). All of the samples were treated using an endotoxin removal kit (Thermo Fisher Scientific) and sterile filtered through a 0.22-µm filter before being aliquoted for immunization studies and stored at −80 °C.
The ternary complex of FlgGEK was produced by co-purifying FlgG, FlgE and FlgK. In brief, supernatants of the three proteins were mixed in a 1:1:1 ratio after lysis and centrifugation, before purification by Ni2+-affinity and S-200 size-exchange chromatography and subsequent endotoxin removal, filtration and freezing, as described above.
dmLT was provided by PATH (Acknowledgements and Data Availability) in 1× PBS supplemented with 0.05% Tween-20 (0.6 mg ml−1).
Animals and study design
Male and female C57BL/6J mice (Jackson Laboratories, 000664) were used in all studies. Mice were assimilated to the new facility 1 week before immunization. Mice were maintained at Vanderbilt University Medical Center under 12 h–12 h light–dark cycles under an ambient temperature of 23 °C (±3 °C) and 50% humidity (±20%), with ad libitum access to chow pellets and water. These studies were approved by the Institutional Animal Care and Use Committee at Vanderbilt University Medical Center and were performed using protocol M2200087-00. All animals were randomly assigned to experimental groups. Researchers were not blinded to groups throughout the animal experiments to properly monitor individual weight loss and morbidity during C. difficile infection according to institutional euthanasia guidelines.
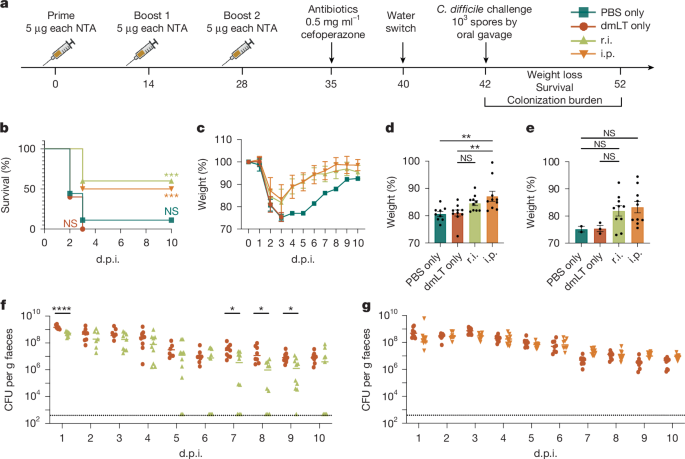
For NTA-cocktail and individual NTA studies, 6-week-old male and female mice were immunized three times over the course of 28 days, with 14 days spanning between injections. Intraperitoneally injected mice received 5 µg of dmLT adjuvant with 5 µg of FlgGEK, C40 peptidase 2, CspC and/or polysaccharide deacetylase in sterile PBS in a total volume of 100 µl per injection. r.i.-treated mice received 25 µg of dmLT adjuvant with 5 µg of FlgGEK, C40 peptidase 2, CspC and/or polysaccharide deacetylase suspended in 200 µl of PBS. Mice were rectally instilled after faecal collection to empty the colon. r.i. occurred under anaesthesia using a sterilized metal ball-end gavage needle that was inserted into the rectum. The vaccine formula was pulsed into the colon, and the rectum was manually squeezed shut for 15 s after administration to prevent leakage, as described previously42. All vaccinations were administered within 2 h of antigen thaw. Faecal and serum samples were obtained before each vaccination and challenge. Mice were challenged 14 days after the final boost, as previously described43,44. Antibiotic treatment was administered by providing 0.5 mg ml−1 cefoperazone in the drinking water ad libitum for 5 days, followed by a 2-day recovery period where normal water was provided before CDI through oral gavage. Two different C. difficile strains were used where indicated: WT R20291 and R20291 ∆A∆B45, both administered at a dose of 1 × 103 spores per mouse. Mice were monitored daily for survival and weight loss. Animal cages were kept the same (left unchanged) for the entirety of the infection. Mice were humanely euthanized when weight loss exceeded 20% of their original body weight. Faecal samples were obtained daily during challenge for CFU enumeration.
For studies to analyse the toxicity of various TcdA and TcdB point mutants, 6-week-old mice were intraperitoneally injected as described above with 5 µg of dmLT and either 1 or 5 µg of the following toxins/toxin combinations: TcdAGTX; TcdB2GTX,L1106K; TcdAGTX + TcdB2GTX,L1106K; TcdB2GTX,L1106K,D1812G; or TcdB2GTX,L1106K + TcdB2GTX,L1106K,D1812G. For combination vaccines with two antigens, 1 or 5 µg of each antigen was injected for a combined total antigenic amount of 2 or 10 µg. The mice were monitored for signs of morbidity and mortality for 7 days after injection.
For studies to optimize the amount of dmLT to include in vaccination, 6-week-old mice were either i.p. injected or rectally instilled with varying amounts of dmLT. Mice were intraperitoneally injected twice over 2 weeks with 0, 0.5, 1, 2.5 or 5 µg of dmLT alongside 5 µg of TcdAGTX or rectally instilled twice over 2 weeks with 0, 10, 15, 20 and 25 µg dmLT with 5 µg of TcdAGTX. Sera and faeces were collected at days 0 (first dose), 14 (second dose), 28, 58 and 88 for enzyme-linked immunosorbent assays (ELISA) analysis of vaccine-induced humoral immune responses.
For studies comparing vaccination of dmLT adjuvant alone to toxin mutants with dmLT and toxin mutants with the NTA cocktail and dmLT, 2 cohorts of 6- and 12-week-old mice were immunized twice with vaccinations spaced 14 days apart. Intraperitoneally injected mice received 1 µg dmLT; 1 µg dmLT with 5 µg each of TcdAGTX, TcdB2GTX,L1106K, TcdB2GTX,L1106K,D1812G; or 1 µg dmLT alongside 5 µg each of TcdAGTX, TcdB2GTX,L1106K, TcdB2GTX,L1106K,D1812G, CspC, C40 peptidase 2 and FlgGEK. r.i.-treated mice received: 15 µg dmLT; 15 µg dmLT with 5 µg each of TcdAGTX, TcdB2GTX,L1106K and TcdB2GTX,L1106K,D1812G; or 15 µg dmLT alongside 5 µg each of TcdAGTX, TcdB2GTX,L1106K, TcdB2GTX,L1106K,D1812G, CspC, C40 peptidase 2 and FlgGEK. Serum and faecal samples were collected as described above. Mice were challenged as stated above with WT C. difficile R20291. Mice were either euthanized for histopathological analysis and flow cytometry analysis on day 3 after infection or were monitored for 10 (6-week-old mice) or 15 (12-week-old mice) days after infection as described above.
For relapsing infection studies, 6 week-old mice were immunized by r.i. with the same experimental groups and same amounts of adjuvant and antigens as noted above. Then, 2 weeks after the final boost, the mice were challenged as stated above with WT C. difficile R20291. Two days after infection, the mice received 0.5 mg ml−1 vancomycin ad libitum in the drinking water for 10 days to clear infection, as described elsewhere46,47. After receiving vancomycin for 10 days, the mice were returned to regular drinking water and monitored for 30 days to document relapsing infection. Stool samples were scored on a 1–5 scale for colour and composition, similar to our previous work48, and were determined as follows: 5, normal, well-formed stool; 4, well-formed, slightly moist or slightly discoloured stool; 3, moist and discoloured stool; 2, soft diarrhoea without wet tail; and 1, wet tail, watery diarrhoea and empty rectum. Animal cages, food and water bottles were changed daily for the entirety of the infection to reduce cross-contamination risk.
For passive transfer studies, 6-week-old donor mice were immunized by rectal instillation with either 15 µg dmLT; or 15 µg of dmLT alongside 5 µg each of C40 peptidase 2 and FlgGEK. Then, 2 weeks after boost, faecal samples were collected from donor mice, as were colonic and caecal contents post-mortem. Faecal and caecal contents were pooled per group and homogenized in 1:1 (w/v) PBS with 2% lysis mix (described above) before centrifugation for 10 min at 10,000g. The resultant supernatant was removed, sterile-filtered with 0.22 µm filters and incubated with anti-mouse IgG MicroBeads (Miltenyi Biotech) for extraction of faecal IgG using an LS Column (Miltenyi Biotech) and MidiMACS magnet system (Miltenyi Biotech). A total of 3.8 mg ml−1 and 3.3 mg ml−1 of faecal IgG were obtained from dmLT-only and C40 peptidase 2- and FlgGEK-vaccinated donors, respectively. Concentrations were obtained on a Nanodrop One C Microvolume UV Spectrophotometer (Thermo Fisher Scientific). Meanwhile, a cohort of 6-week-old recipient mice were r.i.-vaccinated with 15 µg of dmLT alongside 5 µg each of TcdAGTX, TcdB2GTX,L1106K, TcdB2GTX,L1106K,D1812G and CspC. The recipient mice were challenged as stated above with WT C. difficile R20291. At days 1, 4 and 7 after infection, mice received a passive transfer of isolated, sterile-filtered faecal IgG from donor mice either by i.p. (100 µl) or r.i. (200 µl). Recipient mice were monitored for 10 days after infection, as noted above. The animal cages were changed daily for the entirety of the infection.
For studies comparing vaccination formulas with and without the inclusion of CspC, 6-week-old mice were rectally instilled with 15 µg dmLT; 15 µg dmLT alongside 5 µg each of TcdAGTX, TcdB2GTX,L1106K, TcdB2GTX,L1106K,D1812G and CspC; or 15 µg dmLT alongside 5 µg each of TcdAGTX, TcdB2GTX,L1106K, TcdB2GTX,L1106K,D1812G, C40 peptidase 2 and FlgGEK. The mice were challenged as stated above with WT C. difficile R20291. The animals were monitored for 10 days after infection as described above, and the cages were changed daily for the entirety of the infection.
For longevity studies, two cohorts of 6-week-old mice were immunized by r.i. with the same experimental groups and same amounts of adjuvant and antigens as noted above. One cohort of mice was challenged as previously stated with WT C. difficile R20291 60 days after boost, whereas the other was challenged 200 days after boost. Mice were either euthanized for flow cytometry analysis on day 3 after infection or were monitored for 10 days after infection as described above. Animal cages were changed daily for the entirety of both infections.
Antigen-specific antibody measurements
ELISAs were performed. Nunc MaxiSorp 384-Well Plates (Thermo Fisher Scientific) were coated with 30 µl per well of 1 µg ml−1 recombinant TcdA, TcdB2, CspC, C40 peptidase 2, FlgGEK or polysaccharide deacetylase. After overnight incubation at 4 °C, the plates were washed three times with PBS with 0.1% Tween-20 (PBS-T) (100 µl per well per cycle) and replaced with blocking solution (PBS with 0.1% Tween-20 and 2% (w/v) BSA). The plates were then incubated, rocking for 1 h at room temperature. Faeces were homogenized in 1:1 (w/v) PBS with 2% lysis mix (described above) and centrifuged for 10 min at 10,000g. Serum samples were diluted 1:100 in PBS with 2% lysis mix. Plates were washed three times with PBS-T before incubating with 20 µl per well of diluted samples for 2 h, rocking at room temperature. The plates were washed thrice with PBS-T before incubating for an hour with 30 µl per well of horseradish peroxidase (HRP) F(ab’)2-specific goat anti-mouse IgG (Jackson ImmunoResearch; 1:2,000) or goat anti-mouse IgA-HRP (Southern Biotech; 1:2,000) in PBS-T with 2% BSA. Plates were washed four times with PBS-T. Then, 30 µl of TMB substrate reagent (Thermo Fisher Scientific) was added to plates and 30 µl of 2 M sulfuric acid was added after 3 min of substrate development. Plates were recorded at wavelengths of 450 nm using a BioTek Cytation 5 plate reader (Agilent). Faecal antibody titres were normalized to milligram of faeces. Graphs were generated using GraphPad Prism v.10.4.2, and statistical differences between groups were assessed using one-way ANOVA and Tukey’s HSD test.
C. difficile enumeration
C. difficile burden in the stool was quantified by counting CFU from serially diluted stool in 1× PBS (pH 7.4) and plated on taurocholate-cycloserine-cefoxitin-fructose agarose (TCCFA) semi-selective medium49. Faeces was heat treated for 20 min at 75 °C to kill vegetative cells before plating on TCCFA to enumerate spores50. Similarly, bacterial burden in caecal and colonic tissue were quantified by macerating dissected tissues in 1× PBS until slurries were created and plating on TCCFA agar to enumerate total bacterial and spore burdens. Graphs were generated using GraphPad Prism v.10.4.2, and statistical differences between groups were assessed for each day using one-way ANOVA and Tukey’s HSD test.
For PCR analyses, DNA from caecal and colonic tissues (macerated for bacterial burden above) was extracted using the QIAamp PowerFecal Pro DNA Kit (Qiagen). The C. difficile-specific 16S rRNA-encoding gene PCR was used to verify the presence of the bacterium in tissue samples. The PCR set-up was identical to previous published protocols51,52, with template DNA normalized to 50 ng among all of the samples. The following primers were used (ordered from IDT): forward primer 5′-TTGAGCGATTTACTTCGGTAAAGA-3′ (25 nmol, standard desalt purification); reverse primer 5′-CCATCCTGTACTGGCTCACCT-3′ (25 nmol, standard desalt purification)53. PCR products were run on a 1% agarose gel and imaged using a ChemiDoc MP (Bio-Rad).
For qPCR analyses, reactions were set up precisely as outlined elsewhere53, using the same primers as listed above for PCR and TaqMan Fast Advanced Master Mix for qPCR (Thermo Fisher Scientific). Template DNA extracted above from the caecal and colonic tissues were normalized to 50 ng among all samples. Primer amplifications was identified using a C. difficile-specific 16S probe (ordered from IDT): 5′-6-FAM-CGGCGGACGGGTGAGTAACG-MBG-3′ (100 nmol, HPLC purification). Reactions were run on the QuantStudio 6 Flex qPCR system (Thermo Fisher Scientific). Graphs were generated using GraphPad Prism v.10.4.2, and statistical differences between groups were assessed using one-way ANOVA and Tukey’s HSD.
Differential scanning fluorometry
0.1 mg ml−1 each of recombinant WT VPI TcdA, VPI TcdAGTX, WT TcdB2, TcdB2GTX,L1106K and TcdB2GTX,L1106K,D1812G were loaded into glass capillaries and tested using a Tycho differential scanning fluorometer (NanoTemper) according to the manufacturer’s instructions.
Cell rounding assay
Vero–GFP cells54 were seeded in a black, clear-bottom 96-well plate at a concentration of 25,000 cells per well and incubated overnight. Recombinant WT TcdB2, TcdB2GTX, TcdB2L1106K, TcdB2GTX,L1106K, TcdB2GTX,L1106K and TcdB2GTX,L1106K,D1812G were serially diluted tenfold in culture medium, and 100 μl of each sample was added to individual wells in technical duplicate. The plates were statically incubated in a BioTek Cytation 5 plate reader (Agilent) at 37 °C under 5% CO2 and imaged under the bright-field and GFP channels every 45 min at ×20 magnification for 24 h. This experiment was performed twice. The normalized number of rounded cells versus the number of total cells per image was calculated and analysed for concentrations of 1 pM at selected timepoints as previously described54. Graphs were generated using GraphPad Prism v.10.4.2, and statistical differences between groups were assessed using two-way ANOVA and Tukey’s HSD test.
Histopathology
Caeca and colons were fixed in 10% neutral-buffered formalin, dehydrated in graded ethanol series, cleared with xylenes and embedded in paraffin. Tissue blocks were sectioned at a thickness of 5 µm on the HM 335E microtome (Microm) onto Superfrost Plus microscope slides (Thermo Fisher Scientific). To assess histopathology, caecum and colon sections were stained with haematoxylin and eosin (Vector Labs), and conditions were masked for a board-certified gastrointestinal pathologist and a board-certified veterinary pathologist to separately score oedema, inflammation and epithelial damage based on published criteria (n = 5 per treatment)55,56. The averages of the scores from the two pathologists were reported. Histological scores were graphed using GraphPad Prism v.10.4.2, and statistical differences were determined using one-way ANOVA and Tukey’s HSD test. Presented images were captured using a BioTek Cytation 5 automated digital image system (Agilent). Whole slides were imaged at ×10 magnification to a resolution of 0.25 µm px−1.
C. difficile motility assay
WT C. difficile R20291 was grown at 37 °C in BHIS (37 g l−1 brain–heart infusion broth supplemented with 5 g l−1 yeast extract) anaerobic conditions using a COY anaerobic gas chamber (COY Laboratory Products) until mid-log phase. 1 ml of C. difficile was centrifuged at 10,000g for 5 min and the supernatant was removed. The bacterial pellet was either resuspended in 1 ml of 1× PBS (pH 7.4), sterile-filtered dmLT faecal IgG, or sterile-filtered anti-FlgGEK and -C40 peptidase 2 faecal IgG (obtained as noted in the passive transfer mouse experiment, above) and incubated for 30 min. Then, 10 µl of bacterial–faecal IgG mixture was then spotted onto BHIS plates containing 0.3% agar, as previously described57. The plates were incubated at 37 °C for 8 h and room temperature for 16 h under anaerobic conditions. Measurements of the diameter of the widest point of bacterial growth were taken for each spotted colony with a ruler. PBS-incubated C. difficile grew a lawn on BHIS plates, so measurements were taken until the edge of swimming motility, before lawn growth.
Spleen and mesenteric lymph node collection
Spleens were collected, macerated into single-cell suspensions, and filtered using 70 µm cell strainers in 1× Hank’s buffered saline solution (1× HBSS) (Thermo Fisher Scientific). Red blood cells were lysed using ACK lysis buffer (Thermo Fisher Scientific) to obtain a single-cell suspension. Cells were centrifuged and resuspended in 500 µl 1× HBSS, counted and used immediately. Gut-draining mesenteric lymph nodes were collected and processed as described above (without ACK).
Preparations of colons to obtain IELs and LPLs
IEL and LPL fractions were obtained and validated as previously described58,59. In brief, colons were collected, cleaned of fat residue and faeces, and cut open longitudinally before two sequential washes with cold 1× HBSS. The colons were cut into ~0.7 cm chunks and added to 15 ml conical vials with 2 ml of ice-cold HBSS. To these, 5 ml of DTT mix (1× HBSS supplemented with 20 mM HEPES, pH 8.0, 1 mM sodium pyruvate and 1 mM DTT) was added before a 15 min incubation at 37 °C. The conical vials were then shaken by hand for 2 min. The supernatant was transferred to a new 15 ml conical vial containing 5 ml cRPMI-10% FCS (RPMI-GlutaMax supplemented with 10% FBS and 10 mM HEPES, pH 8.0), whereas the tissue was reserved for LPL extraction (below). IELs in the supernatant were further enriched using a Percoll density gradient (Sigma-Aldrich). IELs were centrifuged for 20 min at 4 °C and 650g. The supernatant was aspirated, and IELs were resuspended in 250 µl of 1× PBS supplemented with 0.5% BSA and 1 mM EDTA (constituting PBE buffer). Cells were immediately stained for flow cytometry analyses.
For the LPL compartment, the remaining colonic tissue was kept in the original 15 ml conical vial. Then, 5 ml of EDTA-only mix (1× HBSS supplemented with 20 mM HEPES, pH 8.0, 1 mM sodium pyruvate and 0.5 mM EDTA) was added. The samples were incubated for 10 min at 37 °C. Next, the vials were shaken by hand for 2 min and the supernatant was discarded. Tissue was removed with tweezers and finely minced with scissors into a new 15 ml conical vial containing 3 ml digest mix (1× HBSS supplemented with 20% FBS, 3 mg collagenase D and 0.06 mg DNase I). Samples were then incubated at 37 °C for 30 min. Tubes were shaken by hand for 1 min before pipetting supernatant into new 15 ml conical vials containing 2 ml cRPMI-10% FCS (RPMI-GlutaMax supplemented with 10% FBS and 10 mM HEPES, pH 8.0). LPL cells were centrifuged 20 min at 4 °C and 650g. The supernatant was aspirated, and LPLs were resuspended in 250 µl of 1× PBE. Cells were immediately stained for flow cytometry analyses.
Production of fluorescently labelled recombinant proteins for antigen-specific B cells
Fluorescently labelled recombinant TcdA, TcdB, CspC, C40 peptidase 2, and FlgGEK were prepared by biotinylation using a EZ-Link Sulfo-NHS-LC-Biotinylation kit (Thermo Fisher Scientific, 21435) and conjugated to Streptavidin-linked BV650 (BioLegend, 405231), Alexa Fluor 568 (Thermo Fisher Scientific, S11226), FITC (BioLegend, 405201), APC-Cy7 (BioLegend, 405208) and Alexa Fluor 680 (Thermo Fisher Scientific, S32358). Labelling reactions were calculated to produce a 1:1 ratio of fluorophore:protein. Labelled proteins were flash-frozen in liquid N2 and stored at −80 °C until use to prevent degradation.
Flow cytometry analysis of B and T cells
Single-cell suspensions were incubated with Zombie Near-IR cell viability dye (BioLegend, 423105) for 30 min at room temperature. Cells were washed with 1× PBE, centrifuged at 650g for 10 min and then resuspended in 1× PBE with 5% normal goat serum (60 mg ml−1, Thermo Fisher Scientific) for blocking at room temperature for 30 min. For T cell analysis, cells were stained with anti-CD45 (eFluor 450, 30-F11, 1:600, Thermo Fisher Scientific, 50-112-9409), anti-∆yTCR (PerCP-Cy5.5, GL3, 1:600, BioLegend, 118117), 5-OP-RU tetramer (PE, 1:1,500, NIH Tetramer Core Facility; Data availability), anti-TCRb (Alexa Fluor 594, H57-597, 1:600, BioLegend, 109238), anti-B220 (FITC, RA3-6B2, 1:600, BioLegend, 103205), anti-CD4 (BV570, RM4-5, 1:150, BioLegend, 100541), anti-CD8 (Alexa Fluor 532, 53-6.7, 1:300, Thermo Fisher Scientific, 58-0081-80), anti-CD69 (APC, H1.2F3, 1:300, BioLegend, 104513), and anti-CD103 (PE-Fire 810, QA17A24, 1:300, BioLegend, 156919) for 30 min at room temperature. For B cell analysis, cells were stained with anti-CD45 (eFluor 450, 30-F11, 1:600, Thermo Fisher Scientific, 50-112-9409), anti-B220 (BV480, RA3-6B2, 1:300, BD Biosciences, 565631), anti-CD27 (BV510, LG.3A10, 1:300, BD Biosciences, 563605), anti-CD138 (BV785, 281-2, 1:300, BioLegend, 142534) and the fluorescently labelled recombinant vaccine antigens noted above for 30 min at room temperature. Cells were washed with 1× PBE and centrifuged at 650g for 10 min before fixation in 4% paraformaldehyde fixation for 30 min at room temperature. Cells were then centrifuged, washed with 1× PBE and resuspended in 250 µl of 1× PBE. Flow cytometry data were acquired on a Cytek Aurora Spectral Flow Cytometer (Cytek) and analysed using SpectroFlow Software (v.3.3.0, Cytek). Before flow cytometry analysis, gating of cell populations was determined using Fluorescence Minus One (FMO) controls. FMO controls were prepared by staining replicate cellular samples and beads (Thermo Fisher Scientific, U20250) with all fluorophore-conjugated antibodies in a panel with the exception of one to be analysed for a given control. This accounted for fluorescence spillover and spread across channels and allowed for the precise determination of the positive and negative populations. The gating strategy is provided in the Supplementary Information. Graphs were generated using GraphPad Prism v.10.4.2, and statistical differences between groups were assessed using one-way ANOVA and Tukey’s HSD test.
Priming of naive dendritic cells with antigens and co-culture with TRM cells
BMDCs were isolated from naive C57BL/6J mice and differentiated using GM-CSF (Thermo Fisher Scientific, 315-03-50UG), IL-4 (Thermo Fisher Scientific, 214-14-50UG) and Flt3L (Thermo Fisher Scientific, 250-31L-50UG) as described previously60,61. The day before co-culture with TRM cells, BMDCs were activated with 10 ng ml−1 of lipopolysaccharide (LPS, Thermo Fisher Scientific) and 1 µM of individual antigens or LPS alone. BMDCs were incubated with antigens for 24 h at 37 °C and 5% CO2.
On the day of co-culture, colons from mice vaccinated by i.p. or r.i. with dmLT only or dmLT, toxins and NTAs were collected (n = 3 per group). LPLs were extracted from colons as stated above, pooled within groups, and stained for T cell markers. CD4+ and CD8+ TRM cells were flow-sorted using the Cytek Aurora CS Cell Sorter (Cytek) following the gating strategy provided in the Supplementary Information.
BMDCs were resuspended, washed to remove LPS and growth factors and seeded into 12-well tissue culture plates (Thermo Fisher Scientific) at 5,000 cells per well in 2 ml medium. Based on the counts obtained from the cell sorter, 1,000 CD4+ or CD8+ TRM cells were added to each well of DCs. Cells were incubated together at 37 °C for 72 h before 1 ml of co-culture supernatant was removed and flash-frozen at −80 °C for liquid bead cytokine analysis.
Liquid bead cytokine array
Two custom 12-plex Mouse Luminex Discovery Assay kits (BioTechne) were used to quantify IL-7, IL-12 p70, IL-1β/IL-1F2, IL-2, IL-4, IL-5, IL-6, IL-10, IL-13, IL-17/IL-17a, IFNγ and IL-1α/IL-1F1. Assays were run in triplicate according to manufacturer’s protocol. Acquisition was on a Luminex FlexMap 3D (Luminex). Data were analysed using Millipore Belysa (v.1.0.19) using a four-parameter logistic regression model for calculating concentrations from each standard curve. Graphs were generated using GraphPad Prism v.10.4.2, and statistical differences between groups were assessed using one-way ANOVA and Tukey’s HSD test.
Statistics and reproducibility
Data are presented as mean ± s.e.m. where applicable. Quantitative variables were tested for normal distribution using D’Agostino–Pearson normality tests. If normality was not indicated, then nonparametric statistical tests were used. Statistical tests, parametric or nonparametric, are listed in the figure legends for each experiment and in the corresponding Methods section. Sample variances were also similar between groups unless otherwise mentioned. The range of n within experiments varies based on when samples are taken; for example, lower n values on days 2–3 after infection may be due to animals succumbing to disease or sickness, resulting in an inability to provide faecal samples. Graph schematics were generated using GraphPad Prism v.10.4.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.