Health monitoring and sampling
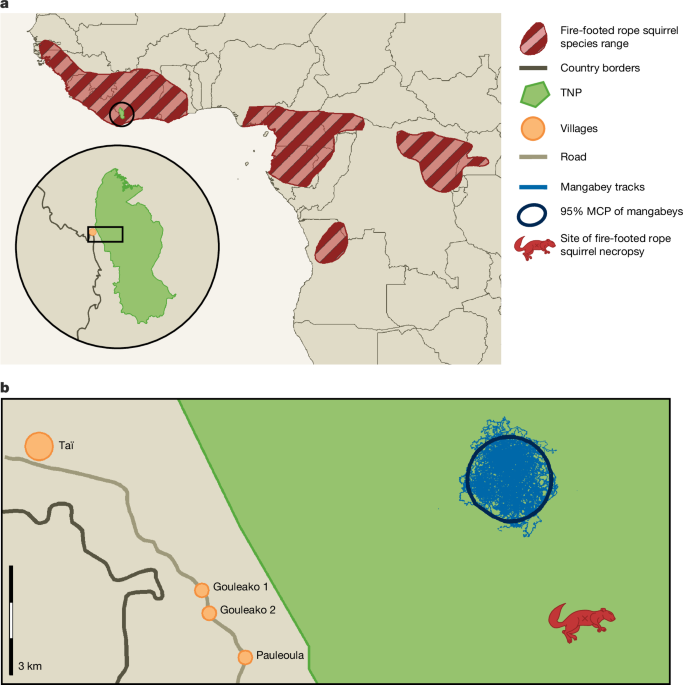
TNP is the largest remaining primary rainforest in Western Africa. Its wild populations of NHP have been studied by the TCP since 1979 (ref. 34). TCP established a veterinary programme from 2001 onwards13. This veterinary programme conducts wildlife mortality surveillance and health monitoring of the four neighbouring groups of chimpanzees and one group of sooty mangabeys that are habituated to human observers. The mangabey group (named the Audrenisrou group) was habituated in November 2012 (ref. 35), and at the time of the outbreak consisted of about 80 individuals. The habituated groups are followed daily by trained field assistants and research staff. Names are given to each habituated individual. Newborns are given a temporary name indicating that they are the infant (BB) of a certain individual (for example, BB-Atacama indicates the newborn of Atacama). Behavioural data, as well as faecal and urine samples, are routinely collected from all the adults of the group. Faecal samples are collected with a plastic spatula right after defecation occurs and stored in 2-ml cryotubes. Urine is collected with fine Pasteur pipettes from underlying vegetation as soon as the animals urinate from a higher position, and is stored in 2-ml cryotubes. These samples are preserved in liquid nitrogen in the field, transported to Germany in dry ice and then stored at −80 °C until further analysis. When clinical signs are observed in the groups, observations and sampling are intensified. During this MPXV outbreak faecal samples were collected in both dry 2-ml cryotubes and in cryotubes containing nucleic acid preserving (NAP) buffer from most individuals of the group belonging to all age categories (infants, juveniles, subadults and adults), and from both symptomatic and asymptomatic individuals. Faecal samples are difficult to collect from infants, therefore the number of these samples is lower than other age classes. It is also important to note that in 2022 a substantial number of male juveniles immigrated to the Audrenisrou group, and many births occurred, leading to an increase in the total population to 80 individuals. To obtain an overview of viral DNA shedding in the mangabey group, we tested faecal samples from three key time periods: from 4 months before the first observations of clinical signs (1 October 2022 to 26 January 2023), during the outbreak (27 January 2023 to 26 April 2023) and up to 4 months after the last symptoms were observed (27 April 2023 to 24 August 2023). A total of 444 faecal samples were tested for MPXV, including those from just before (n = 114), during (n = 170) and after (n = 89) the mpox outbreak in the mangabey group. Details are provided in Supplementary Table 2. The veterinary team of TCP also performs necropsies on all animals found dead in the research area. Necropsies are done by trained veterinarians wearing full personal protective equipment. All used materials are incinerated or disinfected with 1% sodium hypochlorite solution and the carcasses are buried, according to the WHO guidelines. Samples are collected from all inner organs when carcass decomposition is not too advanced and stored in 2-ml cryotubes, both empty and filled with NAP buffer. The cryotubes are then preserved in liquid nitrogen in the field, transported to Germany in dry ice and stored at −80 °C until further analysis. In this study, we included 88 necropsy samples from 23 carcasses representing 11 species (and 4 species for which taxonomic assignment was not possible) collected between 2019 and 2024. Further details are provided in Supplementary Tables 1 and 5.
Trapping of small terrestrial mammals inside and around TNP
Rodents and shrews were trapped using Sherman, Havahart-style or 0.5-m cage traps and were anaesthetized using a combination of ketamine (mouse dose 50 mg kg−1, rats dose 35 mg kg−1; Medistar) and xylazine (mouse dose 5 mg kg−1, rat dose 3.5 mg kg−1; WDT) intramuscularly. After being anaesthetized, the animals were measured, weighed and sampled. After sampling, the animals were marked and placed in an individual box until full recovery was observed. Anaesthetized animals were monitored closely and in some cases antisedan (5 mg kg−1; Vetoquinol) was administered intramuscularly to facilitate recovery. Saline solution was sometimes applied as a subcutaneous infusion to prevent dehydration and applied on the eyes to prevent from drying. After recovery, the animals were released where they were caught. From July to November 2021 and March to April 2023, 173 rodents were trapped in the territory of the mangabey group. From these two field missions, we collected 167 oral swabs, 167 rectal swabs, 23 nasal swabs, 133 faecal samples and 39 samples from skin lesions. Eight individuals died in the trap or succumbed to anaesthetization, and one was euthanized because of signs of extreme weakness. In these cases, necropsies were performed and samples were collected from all main organs. All samples were stored in 2-ml cryotubes dry or with NAP buffer, frozen in liquid nitrogen in the field, transported in dry ice to Germany and then kept at −80 °C. From these trapping missions, a total of 553 samples, including different tissues and swabs were tested for OPVs. We also made use of samples originating from a broader initiative aimed at characterizing the biodiversity of small mammals and related pathogens along a gradient spanning from three villages bordering TNP on the west to the pristine forest in the immediate vicinity of the sooty mangabey territory. We set traps along three parallel transects of about 9 km covering distinct environments: (1) anthropic/domestic (inside houses), (2) at the village periphery, (3) at the edge between cultivated fields and the national park and (4) in the pristine forest of the national park. Sampling was performed from July to September 2021 and April to May 2022. In total, 521 rodents and shrews were trapped and sampled (as mentioned above), of which 82 were euthanised and underwent a full necropsy. Oral and rectal swabs were stored in 2-ml tubes with NAP buffer at room temperature until transport to Abidjan where they were stored at −20 °C. Necropsy samples were stored in 2-ml cryotubes and immediately frozen in liquid nitrogen. Samples were transported to Germany on dry ice and then stored at −80 °C (necropsies) or −20 °C (swabs in NAP buffer). From this sample set, we tested 506 oral swabs, 269 rectal swabs and different organs from the 82 necropsies. In toto, 1,011 samples from different tissues and swabs were tested for OPV. Details for the sampled animals are provided in Supplementary Table 4a,b.
DNA extraction and OPV DNA detection
Nucleic acids were extracted from 40 mg of faecal matter using the GeneMATRIX Stool DNA Purification Kit (Roboklon). For the necropsy samples, 20 mg of tissue were used for DNA extraction with the DNeasy Blood and Tissue kit (Qiagen) or QIAamp Viral RNA Mini Kit (Qiagen). Nucleic acids from virus isolates were extracted using the NucleoMag VET Kit (Macherey-Nagel) and with the RNAdvance Tissue Kit (Beckman Coulter). DNA extracted from faecal and necropsy samples (excluding rodents) was tested for MPXV in duplicate using a TaqMan real-time quantitative PCR (qPCR) targeting the G2R locus36. Each PCR reaction had a total volume of 25 µl and included the following components: 5 µl of DNA template, 11.8 µl of nuclease-free water, 2.5 µl of 10× reaction buffer, 2 µl of 50 mM MgCl2, 1 µl of 2.5 mM dUTPs, 1 µl of 10 µM G2R G forward primer (5′-GGAAAATGTAAAGACAACGAATACAG-3′), 1 µl of 10 µM G2R G reverse primer (5′-GCTATCACATAATCTGGAAGCGTA-3′), 0.5 µl of 10 µM G2R G probe (AAGCCGTAATCTATGTTGTCTATCGTGTCC) and 0.2 µl of Platinum Taq polymerase. PCR cycling conditions consisted of an initial denaturation at 95 °C for 6 min, followed by 45 cycles of 95 °C for 5 s and 60 °C for 30 s. Rodent DNA extracts (including the DNA extracts from trapped rodents and necropsies) were tested in duplicate for OPV using a TaqMan real-time PCR targeting the P4A gene37. Each reaction was prepared in a total volume of 25 µl, consisting of 5 µl of DNA template, 12.7 µl of nuclease-free water, 2.5 µl of 10× reaction buffer, 2 µl of 50 mM MgCl2, 1 µl of 2.5 mM dUTPs, 0.75 µl of 10 µM OPV forward primer (TAATACTTCGATTgCTCATCCAGG), 0.75 µl of 10 µM OPV reverse primer (ACTTCTCACAAATGGATTTGAAAATC), 0.1 µl of 10 µM OPV TMgB probe (6FAM-TCCTTTACGTG+A + T + A + A + A + T + C + A + T) and 0.2 µl of Platinum Taq polymerase. PCR cycling conditions were set to an initial denaturation at 95 °C for 10 min and 45 cycles of 95 °C for 15 s and 60 °C for 34 s. Positive extracts were then tested with the MPXV-specific qPCR mentioned above. A confirmatory PCR targeting a 270 base pair (bp) fragment of the haemagglutinin (HA) gene of OPVs38 was performed for all the extracts that had weakly positive results in the MPXV or OPV qPCRs. For this assay, a single reaction had a total volume of 25 µl, containing 5 µl of DNA template, 11.8 µl of nuclease-free water, 2.5 µl of 10× reaction buffer, 2 µl of 50 mM MgCl2, 2 µl of 2.5 mM dUTPs, 0.75 µl of 10 µM OPV.HA-156 forward primer (GGAGCCCAATTCCATTATTC), 0.75 µl of 10 µM OPV.HA-424 reverse primer (gTATTATgTCTATAgTCgATTCACTATCTg) and 0.2 µl of Platinum Taq polymerase. The PCR protocol included an initial denaturation at 95 °C for 5 min, followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 60 s, with a final extension at 72 °C for 7 min and a hold at 4 °C. The PCR products were then visualized by electrophoresis on a 2% agarose gel.
Mammal species identification
For molecular species identification, two PCR systems targeting the mitochondrial genome were used. The first system designed by Geller and colleagues39 targets the CO1 gene. Each reaction contained 2.5 µl of DNA template, 14.8 µl of nuclease-free water, 2.5 µl of 10× reaction buffer, 1 µl of 50 mM MgCl2, 1 µl of 2.5 mM dUTPs, 1 µl of BSA (1 mg ml−1), 1 µl of 10 µM forward primer jgLCO1490 (5′-TITCIACIAAYCAYAARGAYATTGG-3′), 1 µl of 10 µM reverse primer jgHCO2198 (5′-TAIACYTCIGGRTGICCRAARAAYCA-3′) and 0.2 µl of Platinum Taq polymerase. The PCR protocol included an initial denaturation at 94 °C for 2 min, followed by 47 cycles of 95 °C for 30 s, 52 °C for 30 s and 72 °C for 50 s, with a final extension at 72 °C for 2 min and a hold at 8 °C. The PCR products were then visualized by electrophoresis on a 1.5% agarose gel. The second system targets the cytB gene40. Each reaction contained 1 µl of DNA template, 16.25 µl of nuclease-free water, 2.5 µl of 10× reaction buffer, 2 µl of 50 mM MgCl2, 2 µl of 2.5 mM dUTPs, 0.5 µl of 10 µM forward primer CytB-outF (5′-CGAAGCTTGATATGAAAAACCATCGTTG-3′), 0.5 µl of 10 µM reverse primer CytB-inR (5′-AGTGGRTTRGCTGGTGTRTARTTGTC-3′) and 0.25 µl of Platinum Taq polymerase. The PCR protocol included an initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 52 °C for 30 s and 72 °C for 45 s, with a final extension at 72 °C for 10 min and a hold at 8 °C. The PCR products were then visualized by electrophoresis on a 1.5% agarose gel. If a band was visible at the target lengths of the PCRs, the PCR product was Sanger sequenced. After removal of the primer target-regions in Geneious Prime 2025.1.2 (https://www.geneious.com), a query search of the resulting reads to identify the best sequence matches was performed on Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). If molecular identification of species failed, the animals were determined morphologically following Kingdon Field Guide to African Mammals41 and Mammals of Africa (Vol. III)33.
Virus isolation
Virus isolation was attempted from 13 faecal samples (12 from the mangabeys, 1 from the fire-footed rope squirrel), 13 tissue samples and maggots from two necropsies. Skin, lung and spleen were tested for each mangabey necropsy, as well as a maggot from one individual. The squirrel samples tested encompassed skin, lung, spleen, liver, faeces and maggots. The samples were added to cell culture medium with 10% fetal bovine serum supplemented with penicillin/streptomycin (Gibco) and gentamicin/amphotericin (Gibco), bead homogenized on a bead ruptor and incubated overnight at 8 °C. Sample homogenate was filtered through a 0.8-µm pore membrane to remove larger particles and potential contaminating bacteria. The filtrate was added to confluent layers of MA-104 cells and cultivated with the aforementioned antibiotic-supplemented medium in 12.5-cm2 rectangular canted neck cell culture flasks. MA-104 cells originated from the Collection of Cell Lines in Veterinary Medicine, Insel Riems. The cell line has been authenticated by DNA barcoding of the cytochrome b gene, species-specific PCR, PCR targeting the aldolase gene and restriction fragment length polymorphism analysis. The cell line used in this study was not tested for mycoplasma contamination. Cell cultures were passaged after 3 days. If a cytopathic effect was visible, cells were passaged further to increase the viral titre for shotgun sequencing.
Hybridization capture and high-throughput sequencing
Illumina-compatible dual index libraries were generated from up to 1,000 ng of DNA extracts from necropsies and four mangabey faecal samples (details in Supplementary Tables 1, 2 and 5). Faecal samples were selected on the basis of viral copy number. DNA was fragmented in 50 µl of low EDTA-TE buffer using a Covaris ME220 Focused-ultrasonicator (Covaris) set for a target fragment size of 350 bp (settings: treatment duration 45 s, peak power 50, duty factor 20%, 1,000 cycles per burst, average power 10, temperature 20 °C). Libraries were built from the fragmented DNA using the NEBNext Ultra II DNA kit following the manufacturer’s recommendations. After the adaptor ligation, a 300–400-bp size selection using MagSi magnetic beads (Carl Roth) was performed if the input was higher than 50 ng of total DNA. Quantification of the final libraries was performed using the Kapa HiFi Library Quantification Kit (Roche) or the NEBNext Library Quant Kit for Illumina (New England Biolabs). Libraries were stored at −20 °C until further use. All libraries underwent MPXV target enrichment through in-solution hybridization capture with a previously described OPV bait set10. We used myBaits RNA baits following the myBaits sequence enrichment for targeted sequencing protocol (v.5.0; Daicel Arbor Biosciences) and applied two successive rounds of overnight (16–24 h) hybridization capture. Also, one round of overnight hybridization capture at 65 °C targeting the mitochondrial genome of rodents was performed on a library of the squirrel spleen. To design these custom baits, all complete mitochondrial genomes of rodents available on GenBank were accessed and redundancies were reduced by clustering genomes using CD-HIT30 at a minimum of 88% sequence identity. Final bait design was based on the resulting 239 accession numbers. For capture, only a quarter of the recommended bait quantity was used. Following each round of capture, the hybridized library pools were amplified using the KAPA HotStart Library Amplification Kit (Roche) to obtain a minimum of 200 ng total DNA per library pool. After final quantification using the Kapa HiFi Library Quantification Kit (Roche) or the NEBNext Library Quant Kit for Illumina, the enriched pools were diluted to the recommended concentrations. Sequencing was performed on a MiniSeq platform (Illumina) using the v.3 chemistry (MiniSeq High output Kit for 75 or 150 cycles). For whole-genome sequencing of MPXV from cell cultures, we generated libraries from isolates from two mangabey skin samples and from the squirrel lung using the Rapid-Barcoding Kit v.14 (Oxford Nanopore Technologies) and sequenced them directly on a PromethION 2 solo platform (Oxford Nanopore Technologies) using R10.4.1 PromethION flow cells. Basecaller v.4.3.0 was set to super-accurate basecalling v.4.3.0, 400 bp.
Sequencing data analyses
Reads from different tissues of the same individual were merged to improve viral genome coverage. Raw sequencing reads were quality-filtered using trimmomatic v.0.39 (ref. 42) using the settings: LEADING:30 TRAILING:30 SLIDINGWINDOW:4:30 MINLEN:30. Filtered reads were then mapped to the most recent MPXV genome from TNP (GenBank accession number MN346702) using BWA MEM v.0.7.17-r1188 (ref. 43). Mapped reads were sorted and duplicates removed using SortSam and MarkDuplicates by Picard v.2.13.3 (http://broadinstitute.github.io/picard/). In parallel, paired reads were mapped to the reference genome in Geneious Prime 2023.1.2 (https://www.geneious.com) using default settings to improve coverage of inverted terminal repeat regions of the MPXV genome. Consensus sequences were generated from the reference-based mapping pipeline and the Geneious mapper and checked manually for concordance. Criteria for consensus calling were set to a minimum unique read depth threshold of 20% and a 95% nucleotide frequency in the reads. If a nucleotide at any given position in the genome was found at a frequency less than 95%, an ambiguous base would be automatically called. For the faecal sample for which we obtained good coverage, but lower than the necropsy samples, consensus-calling criteria were set to a minimum unique read depth of 5% and a 50% nucleotide frequency in the reads. For remaining samples in which we obtained a shallow coverage, mapped reads were visually inspected in Geneious but no consensus sequence was called because of their low quality. Ambiguous bases and regions with a difficult read assembly (tandem repeats) were manually checked in consensus sequences of high-quality genomes. Nearly complete viral genomes (excluding the inverted terminal repeats) used for phylogenetic analyses were assembled from the reference-based mapping. The complete mitochondrial genome of the squirrel was de novo assembled from quality-filtered reads using SPAdes v.3.13.0 (ref. 44). Oxford Nanopore reads were quality trimmed using BBDuk Trimmer v.1.0 with the following settings: qtrim=rl trimq=6 minlength=50 ordered=t qin=33 (BBMap—Bushnell B.—sourceforge.net/projects/bbmap) and de novo assembled using Flye v.2.9.2 (ref. 45) (flags –nano-hq; –genome-size 200k). The entire dataset was then remapped against the initially generated sequence through Minimap2 v.2.17 (ref. 46) (ONT mode; including secondary alignments; maximum secondary alignments per read = 5; minimum secondary to primary alignment ratio = 0.8). Owing to data protection rules, all reads that could potentially be of human origin were removed before submission to the European Nucleotide Archive (BBDuk Trimmer v.1.0, mincovfraction=0.66, ref=GCF_000001405.40_GRCh38.p14_genomic.fna).
Phylogenetic analyses
A dataset representing the current known MPXV clade IIa diversity was assembled from publicly available data on GenBank and GISAID47. For identical sequences originating from the same outbreak only one representative genome was selected. We also included partial genomes from Côte d’Ivoire and Liberia from 2024. After evaluating the Côte d’Ivoire sequences through Nextclade v.3.12.036 (ref. 48) quality control (https://clades.nextstrain.org/), we identified three high-quality genomes, which we included in our analysis. Furthermore, two genomes of lower quality were added because of their origin in geographic proximity to TNP. This dataset plus one representative MPXV genome per species from the TNP 2022/2023 outbreak (n = 28) were aligned using MAFFT v.7.505n49. We used Squirrel v.1.2.2 (https://github.com/aineniamh/squirrel) to generate a masked alignment of 197,211 positions. We used this alignment to reconstruct a maximum-likelihood phylogeny using IQ-TREE v.2.1.4b50,51. Branch robustness was assessed by Shimodaira–Hasegawa-like approximate likelihood ratio tests52. We ran a regression of root-to-tip distances versus time and identified the best-fitting root of the resulting tree using TempEst v.1.5.3 (ref. 53) (Supplementary Fig. 9). For molecular clock analyses, we explored more finely the temporal signal in the tree using Phylostems54. The strongest temporal single was detected for a subtree of 24 sequences (Rsq = 0.71; Supplementary Table 7). We used this reduced dataset for further analyses with BEAST v.1.10.5, under strict and uncorrelated log-normal relaxed clock models55. We first validated the presence of a temporal signal with the Bayesian estimation of temporal signal (BETS) approach56. To do so, we compared marginal likelihoods estimates (MLE) of clock models with or without tip dates (in the second case, all tips are assumed to be contemporaneous). We ran several chains of all models and checked their mixing and convergence, as well as sufficient effective sample sizes of model parameters using Tracer v.1.7.2 (ref. 57) We found that the heterochronous model was decisively better than the isochronous one for both the strict (MLEhetero: −256,462.8 versus MLEiso: −256,505.1) and relaxed clock model (−258,458.7 versus −256,471.0), supporting the existence of a temporal signal in both cases and a better performance of the relaxed clock model. We summarized the posterior set of trees under the form of a maximum clade credibility tree using TreeAnnotator v.1.10.5 (distributed with BEAST). All trees were further visualized and edited in iTOL v.7.1 (https://itol.embl.de/).
Diet analysis
The mangabey’s diet was analysed using a metabarcoding approach. The faecal samples used in this particular study (n = 78) were collected just before the mpox outbreak started in the mangabey group (1 October 2022–30 December 2022). We used the Tagsteady protocol58. A first PCR assay targeting a 130 bp fragment of the 16S mitochondrial DNA was performed with tagged 16S mam1 (5′-CGGTTGGGGTGACCTCGGA-3′) and 16S mam2 (5′-GCTGTTATCCCTAGGGTAACT-3′) primers to identify each sample individually. This PCR was performed with the addition of human blocking primer (16Smam_blkhum 5′-CGGTTGGGGCGACCTCGGAGCAGAACCC-3′) to reduce the amplification of contaminant human DNA. A total volume of 25 µl was used for each reaction, which included 2 µl of DNA template. The cycling parameters were set as follows: 95 °C for 10 min, 35 cycles of 95 °C for 12 s, 59 °C for 30 s, 70 °C for 25 s and 72 °C for 7 min. Subsequently, we generated three pools comprising all positive samples together with the positive and negative controls from the same PCR plate (three plates with one replicate each; 262 amplicons in total). After end repair, we indexed the pools by ligation with Illumina full-length Y-adaptors that carried dual matching indexes (P5–P7). The indexed pools were sequenced on an Illumina iSeq 100 System. To analyse the resulting reads, we first assembled a reference database from the EMBL collection of vertebrate sequences (downloaded on 10 July 2024) on which we performed an in silico PCR with the OBItools (v.3.0.1b21) ecopcr command, allowing up to three mismatches between the primer and the reference sequences. We sorted the reads generated from the diet analysis to their respective PCR replicate using their 5′ nucleotide tags using OBItools and removed primer sequences. Paired-end reads were then merged using the OBItools Illuminapairedend command keeping only reads with an alignment quality score of more than 0.8 and a length more than 80 bp. Sequence variants were then collapsed with the obiclean command, but retaining a count of their appearance in each PCR replicate. We then compared the resulting aligned reads with our reference database to try to assign taxons by using the OBItools ecotag command. To consider a wildlife species detection event genuine, we required that at least two of the three replicates contained at least two times the maximum number of reads assigned a taxonomy in the negative controls. We also used Geneious to competitively map the trimmed reads to the mitogenome we generated from the squirrel spleen, as well as a human (NC_012920), chimpanzee (KU308547) and mangabey (NC_028592) mitogenomes. To assess whether this short 16S fragment contained sufficient variation for unambiguous taxonomic assignment, we compared it to all publicly available 16S sequences from squirrel genera known to occur in Côte d’Ivoire. The fragment clearly distinguished fire-footed rope squirrels from all other genera and species (Supplementary Information and Supplementary Fig. 10).
Re-analyses of fly data
To investigate the distribution of fire-footed rope squirrels and sooty mangabeys along a local ecological gradient, we reanalysed a recently published mammal metabarcoding dataset derived from fly DNA31. To do this, we applied the same bioinformatics approaches used for faecal diet analyses to the 100 datasets (25 from the forest, 50 from the edge and 25 from villages) produced by this study (https://doi.org/10.5281/zenodo.7688126).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.