Cell lines and culture methods
All cell lines were cultured in a humidified incubator under normoxia at 37 °C with 5% CO2. Cell lines were validated by STR analysis and were routinely screened and tested negative for mycoplasma.
Mouse cell lines
Immortalized mNSCs were provided by R.J.G.62. RCAS-TVA control (NS-1, NS-2 and NS-3) cell lines or mouse tumour cell lines with the ZFTA–RELA fusion (H-57, H-41 and H-59)12 were provided by E.C.H. Mouse cells were cultured in neurobasal medium (Gibco, 21103049) with 0.2% heparin solution (StemCell Technologies, 07980), 20 ng μl–1 EGF (Shenandoah, 100-26) and 20 ng μl−1 FGF-basic 154aa (Shenandoah, 100-146).
Patient-derived cell lines
EP1NS (ZFTA–RELA+) cells were obtained from T. Milde63. EPD-210 (PFA) cells were obtained from the Brain Tumour Resource Laboratory, Fred Hutchinson Cancer Research Center. Both cell lines were grown in neurobasal medium without vitamin A (Thermo Fisher, 12587-010) with 0.2% heparin solution, 20 ng μl–1 EGF and 20 ng μl–1 FGF-basic 154aa (Shenandoah, 100-146). ST-1 (ZFTA–MAML3+), ST-2 (ZFTA–RELA+) and ST-4 (ZFTA–RELA+, CDKN2A−/−) cells were provided by K.A.M. The cell lines were cultured in neurobasal medium without vitamin A supplemented with Glutamax (Thermo Fisher Scientific, 35050-061), 200 µg ml–1 human EGF and 4 µg ml–1 human FGF and 0.2% heparin in T-75 flasks coated with poly-l-ornithine solution (Sigma-Aldrich, P4957) and laminin from Engelbreth–Holm–Swarm murine sarcoma basement membrane (Sigma-Aldrich, L2020). The CPITT-1 (ZFTA–RELA+) cell line was provided by S.A. and was cultured in neurobasal medium supplemented with 0.2% heparin solution, 20 ng μl–1 EGF and 20 ng μl–1 FGF-basic 154aa. EPN1425 cells were provided by S. Mack and were cultured in DMEM medium (Gibco, 11965092) supplemented with 10% fetal bovine serum (VWR, 89510-186) and 200 mM l-glutamine (Thermo Fisher Scientific, A2916801). MAF-1329 (ZFTA–RELA+) and MAF-811 (PFA) cell lines were provided by A.G. and N.K.F. and were cultured in Opti-MEM media (Gibco, 31985070) supplemented with 10% fetal bovine serum and 200 mM l-glutamine. All cell lines were cultured in media supplemented with penicillin–streptomycin (10,000 U ml–1) (Thermo Fisher Scientific, 115140122) and plasmocin prophylactic (InvivoGen, antmpp).
Lentiviral-transduction-mediated gene silencing using shRNA
Isogenic mNSCs were generated by transfecting cells with ZFTA WT, RELA WT, ZFTA–RELA or EV backbone4 using lentiviral particles (SBI LentiStarter 3.0 kit, V060A). The following lentiviral transfection protocol was used to express ZFTA WT, RELA WT, EV or ZFTA–RELA (which contains 200 bp upstream of the initiating start codon) plasmids into immortalized mNSCs. Similarly, the same protocol was used to knockdown genes with shRNAs. First, 2 × 106 HEK293T cells were plated on 100 mm dishes 36–48 h before transfection. A change of medium was performed the next day to a volume of 5 ml antibiotic-free medium. All lentiviruses were prepared using a Lentistarter 3.0 kit (System Biosciences, LV060A-1). In brief, 2 μg transfer DNA (shACOD1), 20 μg pPackH1 mix and 24 μl PureFection reagent were mixed in 500 μl serum-free DMEM medium and incubated at room temperature for 30 min. The mixture was added dropwise to HEK293T cells and gently swirled to distribute. The transfected cells were then incubated at 37 °C and 5% CO2 to produce virus for 48 h. During this period (around 48 h before intended infection), 1 × 106 ZFTA–RELA mNSCs or EP1NS cells were plated in T-75 flasks. Lentiviral particles were then collected and filtered using a 0.45 µm PVDF filter. Next, lentiviral particles were evenly distributed onto target cells. Lentiviral medium was removed after 24 h and replaced with suitable ZFTA–RELA mNSC or EP1NS cell culture medium. After 48 h, the transfected cells were treated with the appropriate antibiotic for selection. EP1NS cells were treated with 2 μg ml–1 puromycin, whereas ZFTA–RELA mNSCs were selected using 15 μg ml–1 blasticidin. Lentiviral plasmids used for shRNA-mediated knockdown are as follows: shAcod1 (mouse, access ID: NM_008392.1) (Gentarget) and shACOD1 (human) (Horizon Discovery). The following sequences were targeted to knockdown Acod1 in ZFTA–RELA mNSCs: shAcod1-1 (GAGAGCTTTGCTGGTATGATT) and shAcod1-2 (GAGGCATTGGCTATTGCTGTT).
The following sequences were targeted for knocking down ZFTA–RELA in EP1NS (ZFTA–RELA+) cells. The following ZFTA–RELA shRNAs were custom-designed and obtained from Gentarget: shZFTA Fus-1, GCTTGCCCGCCCAAGGGCCCA; shZFTA Fus-2, AGGGCCCAGAACTGTTCCCCC; and shZFTA Fus-3, CAGAACTGTTCCCCCTCATCT.
Human ACOD1 lentiviral cDNA (NM_001258406) and scrambled vector controls were purchased from Origene (SKU RC232825L4).
Snapshot metabolomics
To obtain a comprehensive overview of metabolites, a total of 2 million cells were cultivated in T-75 flasks and incubated for 24 h. Before metabolite collection, a complete change of medium was performed 2 h before the samples were collected. Our metabolomic analysis involved liquid chromatography with tandem mass spectrometry (LC–MS/MS) per a previously described method64. We used an Agilent 1290 UHPLC and 6490 Triple Quadrupole (QqQ) mass spectrometer (LC–MS) for label-free targeted metabolomic analysis. Agilent MassHunter Optimizer and Workstation software LC–MS data acquisition for 6400 series QqQ B.08.00 were used for standard optimization and data acquisition.
For each multiple reaction monitoring (MRM) transition, a retention time with a 1-min left delta and right delta was assigned. Additional parameters included a mass extraction window of 0.05 Da on both sides of the extracted m/z, Agile2 integrator algorithm, peak filter of 100 counts, noise algorithm RMS, noise SD multiplier of 5 min, S/N of 3, accuracy max of 20% max %Dev and quadratic/cubic Savitzky–Golay smoothing algorithm with a smoothing function width of 14 and a Gaussian width of 5.
In reversed-phase liquid chromatography (RPLC), we used a Waters Acquity UPLC BEH TSS C18 column (2.1 × 100 mm, 1.7 μm) in the positive ionization mode. Mobile phase A consisted of 0.5 mM NH4F and 0.1% formic acid in water, whereas mobile phase B contained 0.1% formic acid in acetonitrile. The gradient program involved initially holding mobile phase B at 1% for 1.5 min, followed by an increase to 80% over 15 min, further increasing to 99% over 17 min and holding for 2 min before returning to the initial condition and holding for 10 min.
For hydrophilic interaction liquid chromatography (HILIC), we used a Waters Acquity UPLC BEH amide column (2.1 × 100 mm, 1.7 μm) in the negative ionization mode. Mobile phase A consisted of 20 mM ammonium acetate in water at pH 9.6, whereas mobile phase B comprised acetonitrile. The gradient program involved initially holding mobile phase B at 85% for 1 min, followed by a decrease to 65% over 12 min, further decreasing to 40% over 15 min and holding for 5 min before returning to the initial condition and holding for 10 min.
Both columns were maintained at a temperature of 40 °C, and each sample (3 μl) was injected into the LC–MS system with a flow rate of 0.2 ml min–1. Calibration was performed using an Agilent ESI-low concentration tuning mix. Optimization was carried out on a 6490 QqQ in either the RPLC-positive or HILIC-negative mode for each of the 245 standard compounds (215 compounds for RPLC-positive and 217 compounds for HILIC-negative).
Immunoblotting
Cells were lysed in 1× RIPA buffer containing 1× protease (100× stock, Sigma Aldrich, P8340) and 1× phosphatase inhibitors (100× stock, Sigma Aldrich, P5726). Protein concentrations were quantified using colorimetric bicinchoninic acid assay (BCA) (Pierce BCA protein assay, 23227). Equal amounts of total protein from cell lysate or histone extracts were loaded on 4–15% Mini-Protean TGX precast gels (Bio-Rad, 3450027). Proteins were transferred to PVDF membranes using a Bio-Rad Trans-Blot Turbo transfer system (Bio-Rad, 1704150). Membranes were blocked with 5% bovine serum albumin (BSA) dissolved in TBST (TBS buffer containing 0.1% Tween-20) and incubated in primary antibody diluted in 5% BSA at 4 °C overnight. The following antibodies were used in the immunoblotting experiments: RELA (Cell Signaling Technology, 8242, 1:1,000); GAPDH (Cell Signaling Technology, 2118, 1:10,000); vinculin (Sigma Aldrich, V9264, 1:40,000); ACOD1-human (Abcam, ab222411, 1:1,000 and Novus Biologicals, NBP3-06244, 1:1,000); ACOD1-mouse (Cell Signaling Technology, 17805, 1:1,000); ZFTA (C11orf95) (VWR, 89379-010, AP11349B, 1:1,000); RFP (Abcam, Ab124754, 1:1,000); MAML3 (Invitrogen, PA5-13678, 1:1,000); SLC1A5 (Cell Signaling Technology, 5345, 1:1,000); GLS-human (Cell Signaling Technology, 49363, 1:1,000); GLS-mouse (Invitrogen, PA5-35365, 1:1,000); MYC (Abcam, 32072, 1:1,000); PTEN (Cell Signaling Technology, 9559, 1:1,000); pAKT (S473) (Cell Signaling Technology, 9271, 1:1,000); AKT (Cell Signaling Technology, 4056, 1:1,000); pS6RP (S235/236) (Cell Signaling Technology, 4858, 1:1,000); S6RP (Cell Signaling Technology, 2217, 1:1,000); pGSK3α/β (Cell Signaling Technology, 9331, 1:1,000); GSK3α/β (Cell Signaling Technology, 5676, 1:1,000); H3K4me3 (Cell Signaling Technology, 9751, 1:1,000); H3K9me3 (Cell Signaling Technology, 13969, 1:1,000); H3K27Ac (Cell Signaling Technology, 8173, 1:1,000); H3K27me3 (EMD Millipore, 07-449, 1:1,000); and total H3 (Cell Signaling Technology, 3638, 1:5,000). Next, membranes were washed with TBST three times. Membranes were incubated with species-matched secondary antibodies (1:5,000), goat-anti-mouse (Bio-Rad, 1706516) or goat anti-rabbit (Bio-Rad, 1706515) conjugated to horseradish peroxidase (HRP) for 2 h at room temperature and washed with TBST 3 times (about 5 min each) and TBS one time (around 10 min). Immunoreactivity was detected using Pierce ECL Western blotting substrate (Thermo Fisher Scientific, 32106). Tumour tissues from mice were processed for immunoblotting analysis after lysing red blood cells with RBC lysis buffer (Miltenyi Biotech, 130-094-183). Uncropped and unprocessed scans of all the immunoblots in the study are provided in Supplementary Fig. 1.
IHC analysis
IHC was conducted on ZFTA–RELA+ and control ependymoma patient-derived samples and on mouse tumours using established procedures. The patient tumours were classified by DNA methylation and fusions were determined by RNA-seq. For IHC of metastatic spinal cord, the vertebral column was dissected and decalcified before haematoxylin and eosin staining. Immunohistochemical staining was performed using either a Discovery XT processor from Ventana Medical Systems or a Leica Bond automated staining processor from Leica Biosystems. Each tissue section was blocked with a mixture of 10% normal goat serum and 2% BSA in PBS for 30 min. Subsequently, a rabbit monoclonal anti-ACOD1 antibody (1:200, Abcam, ab238580) or anti-PTEN (1:200, Abcam, ab170941), H3K27me3 (EMD Millipore, 07-449, 1:150), Ki-67 (Invitrogen, MA5-14520, 1:400) and SLC1A5 (Sigma Aldrich, HPA035240, 0.1 µg ml–1) was applied to each section for 5 h. The tissue sections were then treated with biotinylated goat anti-rabbit IgG (PK6101, Vector Labs) at a dilution of 1:200 for 60 min. Chromogens were detected using a DAB detection kit along with streptavidin–HRP and blocker D to minimize background signal (Ventana Medical Systems) following the manufacturer’s instructions. Afterwards, each section was mounted, dried and visualized using an Aperio Vista scanning system (AperioScanscope Scanner). The accompanying Aperio ImageScope software program was used to view each slide at ×40 magnification. An experimenter, who was unaware of the study design, captured JPEG images from three randomly selected areas of each control-stained and ZFTA–RELA-stained section. To quantify the images, an automated analysis program previously published by our laboratory65 was used. This MatLab-based program uses techniques such as k-means clustering, colour segmentation based on RGB colour differentiation and Otsu’s threshold-based background–foreground separation. It generates a quantitative score by multiplying the extracted pixels with the average intensity for each JPEG image.
Cumate-inducible ZFTA–RELA fusion system
The SPARQ 2 Cumate Switch system (System Biosciences, QM822B-1) was used to design the inducible ZFTA–RELA fusion model. The EV and ZFTA–RELA fusion plasmids were transduced in both mNSCs and in HEK293T cells via lentiviral transfection as described above. Cumate (10,000×) purchased from System Biosciences (PBQM100A-1) was used at 1× concentration for 48 h to induce expression of either EV or the ZFTA–RELA fusion.
KDM5 activity measurement
Assessment of KDM5 enzyme activity was performed using a JARID1A Homogenous Assay kit (BPS Bioscience, 50510-2) following the manufacturer’s instructions in three independent experiments. For enzymatic inhibition, KDM5 enzyme (25ng µl–1) was incubated in quadruplicate with different concentrations (100, 250, 500, 1,000, 2,000, 5,000 or 10,000 µM) of itaconic acid (Sigma-Aldrich, I29204), D-2G (Cayman, 16366) or L-2HG (Cayman, 16367). For enzymatic activation, KDM5 enzyme at different concentrations (1.5, 3.0, 6.5, 12.5 or 25.0 ng µl–1) was incubated in quadruplicate with 2,000 µM αKG (Sigma-Aldrich, 349631), 2,000 µM itaconic acid (Sigma-Aldrich, I29204), 2,000 µM D-2HG (Cayman, 16366) or 1,000 µM L-2HG (Cayman, 16367). Compounds were diluted in either PBS or DMSO, which were also used as negative controls. Assays were read in an Envision plate reader (PerkinElmer) using the AlphaLisa 615/Alphascreen 444 method.
ChIP–seq
ZFTA–RELA mNSCs and EPN1425 cells were seeded at a density of 2 × 106 cells in a 100 mm dish and treated with 10 mM dimethyl citraconate (TCI, C0364) or DMSO for 24–48 h. Following this, the cells were detached and dissociated using Accutase (Corning, 25-058-CI). ChIP experiments were performed per the manufacturer’s protocol using an Ideal ChIP–seq kit for transcription factors (Diagenode). For histone ChIPs, 1 million cells were used per ChIP reaction with 2 µg antibody. In brief, cells were crosslinked for 10 min in a 1% formaldehyde solution, followed by quenching with 1/10 the volume of 1.25 M glycine for 5 min at room temperature. Following this, the cells were lysed and sonicated (Bioruptor Pico, Diagenode) to a desired chromatin size of about 200 bp using the Easy mode cycle. Sheared chromatin was then incubated with the following antibodies overnight at 4 °C: H3K4me3 (CST, 9751S) and H3K27me3 (Millipore, 07-449). ChIP DNA was de-crosslinked and purified the next day using a Diagenode iPure kit V3 following the manufacturer’s protocol. Purified DNA was prepared for sequencing using the manufacturer’s instructions (Illumina). About 1–10 ng ChIP DNA was converted to blunt-ended fragments using T4 DNA polymerase, Escherichia coli DNA polymerase I large fragment (Klenow polymerase) and T4 polynucleotide kinase (New England Biolabs (NEB)). Klenow fragment (3′ to 5′ exo minus; NEB) was used to add a single adenine base to fragment ends, followed by ligation of Illumina adaptors (Quick ligase, NEB). PCR enriched the adaptor-ligated DNA fragments using Illumina Barcode primers and Phusion DNA polymerase (NEB). PCR products were size-selected using 3% NuSieve agarose gels (Lonza), followed by gel extraction using QIAEX II reagents (Qiagen). Quantified libraries were quality-checked using a Bioanalyzer 2100 (Agilent) and sequenced on an Illumina HiSeq 2500 Sequencer (125-nucleotide read length).
Reads were processed as previously described66. To summarize, reads were first processed using Trimmomatic (v.0.39) (settings TruSeq3-PE-2.fa:2:30:10, minlen 50) followed by alignment with bwa (bwa mem, options -5SP -T0, v.0.7.17-r1198-dirty) to the mm10 (GRCm38) genome reference or the hg38 (GRCh38) reference67,68. After alignment, the reads were filtered using MarkDuplicates from Picard and then by a quality score of >20 using SAMtools69. MACS2 was used to call peaks, filtered using bedtools and converted to bigwigs with UCSC wigtoBigwig70,71. Cistrome overlap analysis was performed in R (v.3.6.0) using ChipSeekAnno (v.3.0.0) and ChipSeeker (v.1.29.1)72,73. Enrichment heatmaps were generated using DeepTools74.
ATAC-seq
ATAC-seq was performed as previously described75. In brief, 50,000 mNSC and EPN1425 cells were washed in cold PBS and resuspended in lysis buffer (10 mM Tris HCL, pH 7.4, 10 mM NaCl, 3 mM MgCl2, digitonin, NP-40, Tween-20 and protease and phosphatase inhibitor). This single-cell suspension was incubated on ice for 5 min with gentle mixing by pipetting every 2 min, followed by quenching in resuspension buffer. The lysate was centrifuged at 1,300g for 5 min at 4 °C. Nuclei were resuspended in a 50 µl reaction containing 25 µl of 2× TD buffer and 1 µl Tn5 enzyme for 30 min at 37 °C using a Nextera DNA Library Preparation kit. Samples were immediately purified using a Qiagen minElute column and PCR-amplified with NEBNext High-Fidelity 2× PCR master mix (NEB, M0541L). Optimal PCR cycles were determined by qPCR, and the amplified libraries were further purified using a Qiagen minElute column and SPRI beads (Beckman Coulter). Libraries were quantified and quality checked using a Bioanalyzer 2100 (Agilent).
The ATAC-seq libraries were sequenced on an Illumina HiSeq 2500 platform, using a 2 × 50-nucleotide paired-end read length with a sequence depth of 40–45 million. Sequencing of ATAC-seq libraries generated fastq files, which were initially processed using Trimmomatic (v.0.39) for trimming67. These files were then aligned to the mm10 (GRCm38) mouse genome reference or the hg38 (GRCh38) reference using bwa mem (v.0.7.17-r1198-dirty)67, and the alignments were converted to binary format with SAMtools (v.1.9)68. Next, we eliminated mitochondrial and duplicated reads using SAMtools and Picard MarkDuplicates (v.2.26.0-1-gbaf4d27-SNAPSHOT)69. Peaks in the ATAC-seq data were identified using MACS2 (v.2.1.1.20160309)70. Finally, the conversion of data to bigwig format was accomplished using the UCSC tool wigtoBigwig71. The enrichment heatmaps were generated using DeepTools74.
RNA-seq
RNA was isolated from 1 × 106 cells seeded in 100 mm dishes using Trizol (Invitrogen 15596-026) and treated with DNase (Sigma, 9003-98-9). RNA-seq libraries were prepared according to the Illumina TruSeq protocol and were sequenced on a HiSeq 2000. RNA-seq data generated were aligned to the mouse reference genome using bowtie and analysed using the RSEM software package with default parameters. DEseq2 was used to identify differentially upregulated and downregulated genes76. Differentially enriched pathways were identified using Enrichr (https://maayanlab.cloud/Enrichr/)77.
Generation of the ZFTA–RELA KDM5δ mutant
The following sequences with RFP tags were transfected into mNSCs to derive ZFTA–RELA and ZFTA–RELA KDM5δ cells (shown in Fig. 3 and Extended Data Fig. 5). Putative KDM5 recognition sites in the ZFTA portion of the fusion were identified based on previous studies36,37,39 and mutated as indicated below.
The KDM5A site in ZFTA exon 1, normally CCGCCC, was changed to CCACCA. The KDM5B site in ZFTA exon 2, normally GCACAC, was changed to GCAAAC. In detail, the parts of the sequence involved are highlighted in bold:
ZFTA–RELA unaltered: GATCCCATGGAGCCCGGCGGGGACCACCGTAAGAGCCGGAGCAGCGGCGGCAGGGGCGGCCCCGGGCCAGCAGTGGCCTCGGCACGGGGCCGACGGCTGCCGCCCGCCGGATCGAGCGGCAGCGCGGAGCCAGAGGAAGACGAAGGCGGGCAAGATCTTCAGCTGGAAGGGGGTGCCTTGGGGTCCTGGGGGAGTGCCCCCCTGCCCTCCTCCAGGGCCAGGGGACCAGCATCTTCAGGCAGGAAATATTCAGACCACTGTGAGGCCCGGGCCTCGAGGCCTGGAAAGAGCCGCATCCCTGGCCGTGACCACCGGCGCTACTACCACGACCACTGGCGGCTGGAGTACCTGATGGACTTCAACCCTGCCCGGCACGGCATGGTGTGCATGGTGTGCGGCAGCTCCCTGGCCACCCTCAAGCTCAGCACCATCAAGCGCCACATCCGCCAAAAGCACCCCTACTCCTTGCATTGGAGTCCCCGGGAGAAGGAAGTCATCAGCAACAGCTGGGATGCACACATGGGGCTGGGGGCCTGCGGAGAGGCCGAGGGCCTGGGGGTCCAGGGGGCTGAGGAGGAGGAGGAGGAGGAAGAAGAGGAGGAGGAGGAGGGGGCCGGTGTCCCAGCTTGCCCGCCCAAGGGCCCAG.
ZFTA–RELA KDM5δ: GATCCCATGGAGCCCGGCGGGGACCACCGTAAGAGCCGGAGCAGCGGCGGCAGGGGCGGCCCCGGGCCAGCAGTGGCCTCGGCACGGGGCCGACGGCTGCCACCAGCCGGATCGAGCGGCAGCGCGGAGCCAGAGGAAGACGAAGGCGGGCAAGATCTTCAGCTGGAAGGGGGTGCCTTGGGGTCCTGGGGGAGTGCCCCCCTGCCCTCCTCCAGGGCCAGGGGACCAGCATCTTCAGGCAGGAAATATTCAGACCACTGTGAGGCCCGGGCCTCGAGGCCTGGAAAGAGCCGCATCCCTGGCCGTGACCACCGGCGCTACTACCACGACCACTGGCGGCTGGAGTACCTGATGGACTTCAACCCTGCCCGGCACGGCATGGTGTGCATGGTGTGCGGCAGCTCCCTGGCCACCCTCAAGCTCAGCACCATCAAGCGCCACATCCGCCAAAAGCACCCCTACTCCTTGCATTGGAGTCCCCGGGAGAAGGAAGTCATCAGCAACAGCTGGGATGCAAACATGGGGCTGGGGGCCTGCGGAGAGGCCGAGGGCCTGGGGGTCCAGGGGGCTGAGGAGGAGGAGGAGGAGGAAGAAGAGGAGGAGGAGGAGGGGGCCGGTGTCCCAGCTTGCCCGCCCAAGGGCCCAG.
ChIP–PCR
ZFTA–RELA mNSCs and EPN1425 cells were seeded at a density of 5 × 106 cells in a 100 mm dish and treated with 10 mM dimethyl citraconate (TCI, C0364) or DMSO for 48 h. Chromatin fragmentation was achieved by sonication (QSonica, 800R) and by micrococcal nuclease (Cell Signaling Technologies, 10011) digestion to a final size of 150–300 bp. ChIP was performed using a Simple Chip Enzymatic Chromatin IP kit (Cell Signaling Technologies, 9002) and a Simple Chip Plus Sonication Chromatin IP kit (Cell Signaling Technologies, 56383) as per the manufacturer’s protocols.
Two independent set of primers were designed and used for the ChIP PCR and RT–qPCR experiments shown in Fig. 3h and Extended Data Fig. 5e,f,j. They were custom designed and obtained from Integrated DNA Technologies (IDT). The following ZFTA-based primers were used: ZFTA exon 5 set 1 FWD: 5′-AGAGGAGGACGAAGAGGAC-3′; ZFTA exon 5 set 1 REV: 5′-CCGTCGTAGTCCATCAGGTA-3′; ZFTA exon 5 set 2 FWD: 5′-TGAGGAGCGCCAGACTAT-3′; ZFTA exon 5 set 2 REV: 5′-CACACAGCGCCAGACTT-3′; ZFTA exons 1–2 set 1 FWD: 5′-GAGCCAGAGGAAGACGAAGG-3′; and ZFTA exons 1–2 set 1 REV: 5′-GAAGATGCTGGTCCCCTG-3′. The following RELA-based primers were used: RELA exon 4 set 1 FWD: 5′-ACAGGACCAGGGACAGT-3′; RELA exon 4 set 1 REV: 5′-CTCAGCCTCATAGAAGCCATC-3′; RELA exon 4 set 2 FWD: 5′-CGCATCTCCCTGGTCAC-3′; and RELA exon 4 set 2 REV: 5′-CAT CCC GGC AGT CCT TT-3′. The following mouse primers were used: H3f3a FWD: 5′-ACAAAAGCCGCTCGCAAGAGTG-3′; and H3f3a REV: 5′-TTCTCGCACCAGACGCTGAAAG-3′.
RT–qPCR
RNA was isolated from 1 × 106 cells seeded in 100 mm dishes using Trizol (Invitrogen 15596-026) and treated with DNase (Sigma, 9003-98-9) per the manufacturer’s instructions. qPCR was performed with n = 5 samples of 20 ng mRNA each using a TaqMan RNA-to-CT kit (Applied Biosystems, 4392938). TaqMan probes (Thermo Fisher) targeting mouse Pten (Mm00477208), human PTEN (Hs02621230), human GAPDH (Hs99999905, Thermo Fisher Scientific, 4326317E) and mouse Gapdh (Mm99999915_g1, Thermo Fisher Scientific, 4352339E) were used. The following primers from IDT were used to detect ZFTA–RELA mRNA in patient-derived cell lines (shown in Fig. 1m): fusion set 1 FWD: 5′-GAGGAGGAGGAGGAAGAAGAG-3′; fusion set 1 REV: 5′-GCTGCTCAATGATCTCCACATA-3′; fusion set 2 FWD: 5′-GAAGAAGAGGAGGAGGAGGAG-3′; and fusion set 2 REV: 5′-CTTGGGCTGCTCAATGATCT-3′.
RT–qPCR was conducted using an ABI7900HT Sequence Detection system, and relative expression was calculated using the \({2}^{-\Delta {\Delta }_{\mathrm{CT}}}\) method, with Gapdh (mouse) or TBP (human) as a housekeeping control for normalization.
Glutamine uptake and intracellular glutamine and glutamate measurement
Glutamine uptake and intracellular glutamine levels were estimated using a Glutamine/Glutamate-glo assay kit (Promega, J8022). In brief, 25,000 cells per ml medium per well were seeded in triplicate in a 24-well plate for each time point as indicated. Both cell culture media and cells were collected at 0, 12, 24, 36, 48, 60 and 72 h. Following extraction, intracellular and extracellular glutamine and glutamate were measured by bioluminescence detection for all the time points. The concentrations of glutamine and glutamate were calculated using the standards provided by the manufacturer. The values obtained were used to estimate percentage glutamine uptake and intracellular glutamate to glutamine ratios.
Isotope tracing of radiolabelled glucose and glutamine
For 13C tracing, 2 million cells were plated and cultured using medium supplemented with either 13C-U-labelled glucose (25 mM) or glutamine (2 mM, both from Cambridge Isotope Laboratories, CLM-99-1396-1 and CLM-1822-H, respectively) for 16 h. Cells were collected by centrifugation and the medium was aspirated. The cell pellet was resuspended in 300 μl ice-cold methanol followed by thorough pipetting. Next, 300 μl water containing 1 μg norvaline as internal standard and 600 μl chloroform was added to the microfuge tube. The extracts were vortexed thoroughly for 30 min at 4 °C, followed by centrifugation. The polar layer was dried in a SpeedVac for 3–4 h without heat and stored at −80 °C. For derivatization, 30 μl methoxyamine hydrochloride (Fisher Scientific, PI45950) was added to the dried samples and incubated at 45 °C for 30 min with constant mixing. Next 30 μl MBTSTFA and 1% TBDMCS (Sigma Aldrich, M-108-1243 5×1ML) was added, and samples were incubated at 45 °C for 1 h. Derivatized samples were transferred to GC vials with glass inserts. Samples were analysed using an Agilent 7890 GC equipped with a 30-m HP-5MSUI capillary 1246 column connected to an Agilent 5977B MS in scan mode. In total, a 1–2 μl sample was injected at 270 °C with helium as the carrier gas at 1 ml min–1 flow. The temperature gradient was maintained at 100 °C for 1 min, increased at 3.5 °C min–1 to 255 °C, increased to 320 °C at 15 °C min–1 and held for 3 min (method total time of 52.6 min). MS detector was operated in scan mode (70–600 m/z). Relative metabolite abundances were calculated by normalizing the integrated peak area of ions to the internal standard signal (norvaline) and protein content. For 13C-labelled samples, isotopic correction of raw GC–MS peaks for all reported metabolites was performed using the IsoCorrectoR package (v.1.5.1) available as part of the Bioconductor library (BioC 3.8) and implemented in R (CRAN 3.6.1).
Cell proliferation and viability assays
Proliferation and viability of cells after drug treatment was assessed by trypan blue exclusion assays. Cells were seeded at a density of 2.5 × 104 cells per well in a 24-well plate and were left overnight. They were then treated with vehicle or drug (JHU-083, V-9302, CB-839 or PI-103) at varying concentrations and time points for the indicated time periods, following which they were counted using a Countess III cell counter (Thermo Fisher Scientific). Cell number (y axis) for each cell line was calculated as a percentage of living cells normalized to the untreated or vehicle-treated controls (average number of cells alive in the untreated or vehicle-treated wells).
Chemical compounds
The drugs and chemical compounds used in the study are listed as follows: dimethyl citraconate (Fisher Scientific, C0364-5G), mesaconic acid (Millipore-Sigma, 131040-10G), itaconic acid (Sigma Aldrich, I29204), dimethyl 2-ketoglutarate (Cayman Chemical, 28394), octyl-l-2-hydroxyglutarate (Cayman Chemical, 16367), LPS (Cayman Chemical, 19661), IRG1-IN-1 (MedChem Express, HY-148335), GSK-343 (Cayman Chemical, 14094), CPI-455 (Cayman Chemical, 22127), DON (Cayman Chemical, 17580), JHU-083 (MedChem Express, HY-122218), V-9302 (Selleckchem, S8818), CB-839 (MedChem Express, HY-12248), PI-103 (Selleckchem, S1038), MK-2206 (Cayman Chemical, 11593) and DMSO (Sigma Aldrich, D2650).
Histone extraction
Cells were plated at densities below confluence. Following the specified treatment, cells were rinsed with PBS. The cell pellet resulting from centrifugation was resuspended in 1 ml hypotonic lysis buffer (containing 10 mM Tris HCl pH 8.0, 1 mM KCl, 1.5 mM MgCl2 and protease and phosphatase inhibitors) and incubated for 30 min on a rotator at 4 °C. The pellet was then collected by centrifugation at 10,000g, 4 °C for 10 min. It was subsequently resuspended in 400 μl of 0.4 N H2SO4 and left to incubate overnight on a rotator at 4 °C. After centrifugation, the resulting supernatant was transferred to a new tube, and 132 μl trichloroacetic acid was gradually added. The mixture was then incubated on ice for 30 min. The histone pellet was obtained by centrifugation at 16,000g, 4 °C for 10 min and washed with cold acetone. After another round of centrifugation at 16,000g, 4 °C for 5 min, acetone was removed. The histone pellet was further washed with acetone and subsequently allowed to air dry with the caps open at room temperature for 20 min to eliminate any remaining acetone. Finally, the dried histone pellet was resuspended in an appropriate volume of ddH2O, supplemented with protease inhibitors and kept on ice.
Animal handling and housing conditions
Animal experiments were performed after approval from the University of Michigan Committee on the Use and Care of Animals (PRO00010599) and were conducted as per National Institutes of Health (NIH) guidelines for animal welfare. All animals used in these studies were housed in pathogen-free conditions as per IACUC guidelines with continuous access to both food and water in addition to veterinary care. NOD-SCID-IL2R γ-chain deficient (NSG) mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, 005557) aged 4–6 weeks old were used for all experiments involving subcutaneous or orthotopic injections of ZFTA–RELA mNSCs and for the subcutaneous MAF-1329 and MAF-811 PDX models. IUE models were CD-1 (Charles River, Crl:CD1(ICR), 022) or C57BL/6 animals (The Jackson Laboratory, 003771). For all drug trials, the animals were randomized and sorted into treatment groups in an unbiased manner, ensuring that all cohorts contained equal proportions of male and female animals.
Nes
cre
Acod1
fl/+ models
Acod1fl/fl animals were provided by M. Diamond25. B6.Cg.Tg.(Nes Cre)1KlnJ animals were crossed with Acod1fl/fl animals. The pups were weaned after 21 days and genotyped to establish NescreAcod1fl/+ mice.
Subcutaneous ZFTA–RELA xenograft models
In this study, two subcutaneous xenograft models (mouse and human patient-derived) ZFTA–RELA fusion tumours were established in NSG mice. Xenograft models were generated by injecting 1 × 106 ZFTA–RELA mNSCs on either flank of the animal and tumour volumes were measured once palpable tumours (around 200 mm3) appeared (about 1 week after injection). MAF-1329 (ZFTA–RELA+) and MAF-811 PDXs were provided by A.G. and N.K.F. PDX cells were injected in NSG mice and were allowed to grow. Once the tumours reached the end-point volumes of 1,000–1,500 mm3, they were excised and homogenized into single-cell suspensions using a gentleMACS Octo dissociator (Miltenyi Biotech, 130-096-427), following which they were counted and serially passaged in NSG animals to generate a suitably sized cohort for the trial. For subcutaneous tumours, growth was measured using Vernier calipers, and tumour volumes were calculated using the formula L × W × W/2, where L is the longer dimension and W is the shorter dimension of the tumour. At the end point, the animals were humanely euthanized and perfused before collecting the tumour tissues. Tumour volumes were not allowed to exceed these limits (as per IACUC guidelines) in any of the experiments.
Orthotopic ZFTA–RELA tumour models
Orthotopic models of ZFTA–RELA tumours were established in NSG animals by injecting 2 × 105 ZFTA–RELA bioluminescent mNSCs in the cortex, 3 mm posterior and 2 mm lateral right from the bregma using a stereotaxic apparatus. Before implantation, mice were anaesthetized by intraperitoneal injection of ketamine (90 mg kg–1) and dexmedetomidine (0.6 mg kg–1). Carprofen (5.5 mg kg–1) was used as an analgesic to alleviate pain after surgery. A total volume of 4 μl of cells resuspended in sterile PBS was injected using a Hamilton syringe through a burr hole drilled at the location described. The surgery site was treated with iodine and sealed with a wound clip to avoid infection. The animals were then revived using atipamezole (1.25 mg kg–1).
IUE ZFTA–RELA tumour models
IUE ZFTA–RELA immunocompetent ependymoma animal models were established using previous protocols to generate piggy-bac (PB) ZFTA–RELA plasmids containing the luciferase gene along with the plasmid transposase (PBase)6,10,11. Pregnant CD1 or B6 mice (Charles River) mice at embryonic day 15 were used. IUE was performed by injecting a concentrated mixture of DNA (1 μg μl–1 ZFTA–RELA plasmid) along with 0.05% Fast Green (Sigma) into the lateral ventricle of the embryos using a pulled glass capillary pipette. Electroporation was performed using 5 square pulses (45 V, 50 ms pulses with 950 ms intervals) and applied using a 3 mm tweezer electrode, with the positive electrode directed towards the cortex (BTX/Harvard Bioscience). After the procedure, the embryos were carefully placed back into the abdominal cavity, the incision was sutured, and the female mouse was monitored until she fully recovered.
Pharmacological and radiation treatment
In both the implantation and IUE models, tumour engraftment and establishment was verified by bioluminescence measurements. The flux density was calculated, and treatment regimens were initiated only after tumours were established (predefined as >105 photons s–1). Two independent baseline measurements were recorded for every animal in the trial to ensure tumour growth and to avoid possible technical artefacts. Subsequent bioluminescent readings were normalized to the baseline measurements to calculate the fold change in the signal to assess tumour progression. The glutamine antagonist JHU-083 was dissolved in 10% DMSO and 90% corn oil to be dosed orally at 20 mg kg–1 twice a week. The dual PI3K–mTOR inhibitor was dissolved in DMSO and was i.p. injected every other day during weekdays with scheduled breaks in treatment on the weekend. Dimethyl citraconate was prepared in sterile saline solution was i.v. injected on alternate days for the entire duration of the treatment paradigm. For experiments involving radiotherapy, whole-brain irradiation was performed on mice twice per week (single fraction of 2 Gy per day) for two consecutive weeks.
Bioluminescence measurement and analysis
Successful tumour formation was verified by checking for bioluminescence after injecting the pups with luciferin (15 mg ml–1, GoldBio, LUCK 115144-35-9) using an IVIS spectrum (Perkin Elmer) instrument. Once anaesthetized, a sequence of bioluminescence images was recorded to capture the peak flux intensity value for each animal. This value was then used to normalize to baseline for fold change calculation. In the implantation model in which ZFTA–RELA tumours metastasized to the spinal cord, the fold change in tumour bioluminescence signal was calculated by defining a region of interest around the spinal cord distal and distinct to the head. The end points of the animal trial were defined based on IACUC guidelines and animal welfare was prioritized. Swelling of the head, lack of mobility, loss of weight or appetite, among other parameters, were all considered as key criteria for euthanasia of tumour-bearing animals. Tumour volumes were not allowed to exceed these limits (as per IACUC guidelines) in any of the experiments.
Statistics and reproducibility
The Department of Bioinformatics provided consultation for the statistical analyses conducted. Each figure and its corresponding legend indicate the sample size (n), the specific statistical test performed and the corresponding P values. No data were excluded in analyses of samples or animals. Owing to the rarity of ZFTA–RELA ependymomas, the sample sizes were determined based on the available tumour samples. The figure legends provide the exact n value for each experiment. The data are presented as the mean ± s.d., and the box and whisker plots show the maximum, mean and minimum values. Graphs were generated and statistical analyses were performed using Prism software (v.9.5.1, GraphPad). The data analysis methods included unpaired, two-sided, two-tailed Student’s t-test, one-way or two-way ANOVA followed by post hoc Bonferroni multiple comparison analysis along with Dunnett’s correction, as indicated in each figure. Data in Fig. 1h was analysed using nonparametric Spearman’s correlation with 95% confidence intervals and correlation coefficient (R2) and P values are indicated.
Overall survival data, including data from animal models, were visualized using Kaplan–Meier curves and the association of various factors with overall survival was assessed using the log-rank test. Data were considered significant if P values were below 0.05 (95% CI).
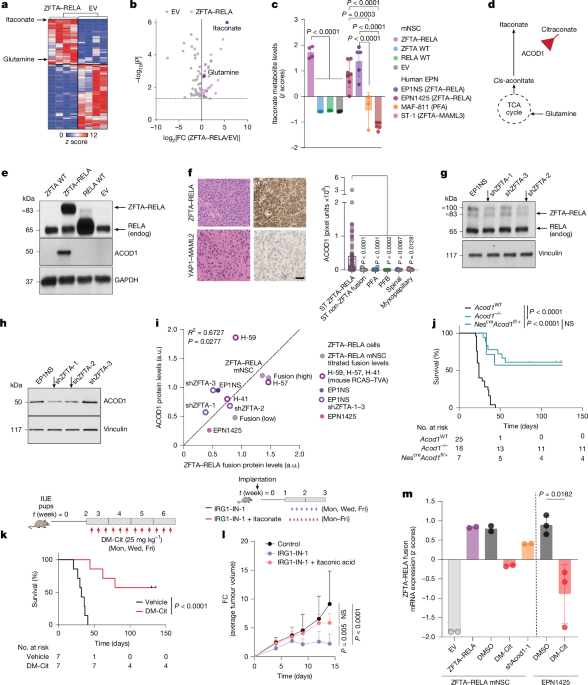
Data shown in the following figures are representative images of independent experiments or samples as indicated. Fig. 1e (n = 3 independent experiments), Fig. 2a (n = 5 independent experiments), Fig. 2g (n = 2 independent experiments), Fig. 2h (n = 2 independent experiments), Fig. 2j (n = 2 independent experiments), Fig. 3b (n = 2 independent experiments), Fig. 3c (n = 2 independent experiments), Fig. 3h (n = 5 independent experiments), Fig. 3l (n = 2 independent experiments), Fig. 5c (n = 2 independent experiments), Fig. 5j (n = 3 independent samples were used for immunohistochemical staining), Extended Data Fig. 1e (n = 2 independent experiments), Extended Data Fig. 1f (ZFTA–RELA (n = 18 independent samples), non-ZFTA fusion (n = 5 independent samples), posterior fossa type A (n = 7 independent samples), posterior fossa type B (n = 2 independent samples), spinal (n = 3 independent samples) and myxopapillary (n = 4 independent samples) of ependymoma tumours from patients). Extended Data Fig. 1h (n = 3 independent experiments), Extended Data Fig. 1i (n = 3 independent experiments), Extended Data Fig. 1l (n = 2 independent experiments), Extended Data Fig. 3a (n = 5 independent experiments), Extended Data Fig. 3b (n = 2 independent experiments), Extended Data Fig. 3c (n = 2 independent experiments), Extended Data Fig. 3d (n = 2 independent experiments), Extended Data Fig. 3e (n = 2 independent experiments), Extended Data Fig. 3f (n = 2 independent experiments), Extended Data Fig. 3h (n = 3 independent experiments), Extended Data Fig. 3i (n = 2 independent experiments), Extended Data Fig. 3j (n = 2 independent experiments), Extended Data Fig. 3k (n = 2 independent experiments), Extended Data Fig. 3l (n = 2 independent experiments), Extended Data Fig. 3m (n = 2 independent experiments), Extended Data Fig. 3n (n = 3 independent experiments), Extended Data Fig. 3o (n = 2 independent experiments), Extended Data Fig. 5c (n = 3 independent experiments), Extended Data Fig. 5d (n = 3 independent experiments), Extended Data Fig. 5e (n = 2 independent experiments), Extended Data Fig. 5f (n = 2 independent experiments), Extended Data Fig. 5n (n = 2 independent experiments), Extended Data Fig. 7b (n = 2 independent experiments), Extended Data Fig. 7c (n = 2 independent experiments), Extended Data Fig. 7d (n = 2 independent experiments), Extended Data Fig. 8c (n = 2 independent experiments), Extended Data Fig. 8d (n = 2 independent experiments), Extended Data Fig. 8h (n = 2 independent experiments), Extended Data Fig. 8i (n = 3 independent samples each for mouse ZFTA–RELA fusion tumours and cortex tissues, n = 18 independent samples for ZFTA–RELA+ tumours from patients and n = 5 independent samples for non-ZFTA (YAP1) fusion tumours from patients), Extended Data Fig. 8m (n = 2 independent experiments), Extended Data Fig. 9c (n = 17 independent samples for ZFTA–RELA+ tumours from patients and n = 5 independent samples for non-ZFTA (YAP1) fusion tumours from patients), Extended Data Fig. 9i (n = 2 independent experiments), Extended Data Fig. 9j (n = 2 independent experiments), Extended Data Fig. 9k (n = 2 independent experiments), Extended Data Fig. 10b (n = 2 independent experiments), Extended Data Fig. 11d (n = 3 independent experiments), Extended Data Fig. 12m (n = 3 independent samples).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.