Mouse strains and mutants
Mice (wild-type C57BL/6J, IMSR, JAX: 000664, RRID: IMSR_JAX:000664; Miwi−/− mutation, MGI: 2182488; and pachytene piRNA mutations listed in Supplementary Table 1) were housed and euthanized according to the guidelines of the Institutional Animal Care and Use Committee of the University of Massachusetts Chan Medical School in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited barrier facility at controlled temperature (22 ± 2 °C), relative humidity (40 ± 15%) and a 12-h day–light cycle. All experimental animals were 2–6 months old6,15,48. sgRNAs (Supplementary Table 1) were designed using a CRISPR design tool (https://www.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE). sgRNAs were transcribed with T7 RNA polymerase and then purified by electrophoresis on 10% denaturing polyacrylamide gels. gRNA (20 ng µl–1) and Cas9 mRNA (50 ng µl–1, TriLink Biotechnologies, L-7206) were injected together into the pronucleus of one-cell C57BL/6 zygotes in M2 medium (Sigma, M7167). After injection, the zygotes were cultured in EmbryoMax Advanced KSOM medium (Sigma, MR-106-D) at 37 °C under 5% CO2 until the blastocyst stage (3.5 days), then transferred into the uterus of pseudopregnant ICR females 2.5 days post coitum. To screen for founders with the mutation, gDNA extracted from tail tissues was analysed by PCR using the primers listed in Supplementary Table 1. All mutant strains were maintained in a C57BL/6 background; all experimental animals were the progeny of at least two backcrosses.
Mouse fertility
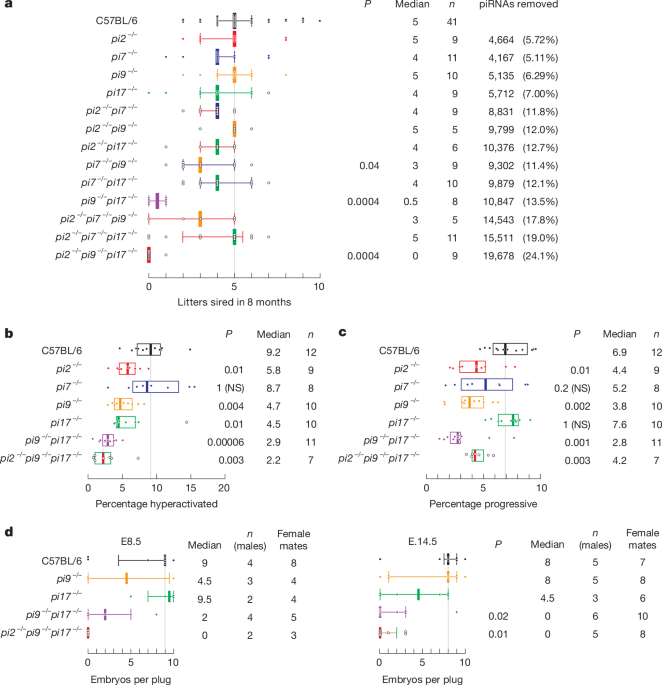
Fertility was measured as previously described6,15,48. In brief, each 2–6-month-old male mouse was continuously housed with one 2–4-month-old C57BL/6 female. For male mice that did not produce pups after 3 months (around 3 cycles), the original female was replaced with a new female and the fertility test continued.
To generate E8.5 or E14.5 embryos, one male mouse was housed with two C57BL/6 females. When a copulatory plug was observed, the female was housed separately until the experiment was completed.
Epididymal sperm count
Sperm counts were obtained as previously described6,15,48. In brief, to quantify sperm abundance, cauda epididymides were collected from mice and placed in PBS. A few incisions were made in the epididymides with scissors to release the sperm, followed by incubation at 37 °C and 5% CO2 for 20 min. A 20 µl aliquot of sperm suspension was diluted in 480 µl of 1% (w/v) paraformaldehyde (PFA) and sperm cells were counted using a Leica DMi8 bright-field microscope equipped with a ×10, NA 0.4 objective.
TUNEL immunohistochemistry
Mouse testes were fixed in Bouin’s solution overnight, washed with 70% ethanol, embedded in paraffin and sectioned at 5 µm thickness. A Click-iT TUNEL Colorimetric IHC Detection kit (Thermo Fisher, C10625) was used to detect DNA breaks according to the manufacturer’s protocol. In brief, testes were fixed and embedded as described above, then were de-paraffinized in three changes of xylene for 5 min each, gradually re-hydrated in 100% (v/v), 95% (v/v) and 70% (v/v) ethanol for 5 min each, and then washed in 1× PBS for 5 min. After pretreating the slides with 20 µg ml–1 proteinase K at room temperature for 15 min, slides were washed with water twice (2 min each). Positive-control slides were treated with 1.0 U Turbo DNase (Thermo Fisher, AM2238) at room temperature for 30 min. Slides were then incubated with TdT reaction buffer containing terminal deoxynucleotidyl transferase in a humidified chamber at 37 °C for 1 h. The reaction was quenched with 2× SSC for 15 min, then washed twice in PBS. Peroxidase activity was quenched in 3% (v/v) H2O2 at room temperature for 5 min. Slides were incubated with biotin azide and copper sulfate in a humidified chamber at 37 °C for 30 min, then stained with peroxidase substrate at room temperature for 10 min. Nuclei were counterstained with haematoxylin I, and the slides were sealed with EcoMount (Biocare Medical, EM897L). Images were captured using a Leica DMi8 bright-field microscope equipped with a ×20 objective with 0.4 NA (HC PL FL L ×20/0.40 CORR PH1, Leica Microbiosystems).
In vitro fertilization and embryo transfer
In vitro fertilization (IVF) was performed as previously described6,15,48,49. In brief, using spermatozoa from caudal epididymis of C57BL/6 or pi9−/−pi17−/− mice, spermatozoa were incubated in complete human tubal fluid medium (101.6 mM NaCl, 4.69 mM KCl, 0.37 mM KH2PO4, 0.2 mM MgSO4·7H2O, 21.4 mM sodium lactate, 0.33 mM sodium pyruvate, 2.78 mM glucose, 25 mM NaHCO3, 2.04 mM CaCl2·2H2O, 0.075 mg ml–1 penicillin-G, 0.05 mg ml–1 streptomycin sulfate, 0.02% (v/v) phenol red and 4 mg ml–1 BSA) with oocytes from B6SJLF1/J mice for 3–4 h at 37 °C with constant 5% O2, 90% N2 and 5% CO2. Oocyte viability and the presence of pronuclei were assessed using a Nikon SMZ-2B (Nikon) dissecting microscope with a ×5, NA 0.6 objective. To observe embryo development, embryos were moved into potassium-supplemented simplex optimized medium (KSOM; 95 mM NaCl, 2.5 mM KCl, 0.35 mM KH2PO4, 0.2 mM MgSO4·7H2O, 10 mM sodium lactate, 0.2 mM sodium pyruvate, 0.2 mM glucose, 25 mM NaHCO3, 1.71 mM CaCl2·2H2O, 1 mM l-glutamine, 0.01 mM EDTA, 0.075 mg ml−1 penicillin-G, 0.05 mg ml–1 streptomycin sulfate, 0.02% (v/v) phenol red and 1 mg ml–1 BSA; Millipore Sigma) after IVF and assessed every 24 h. To measure birth rates, two-cell embryos were transferred to Swiss Webster pseudopregnant females, and fetuses were isolated by caesarean section 18.5 days after embryo transfer. For zona-free IVF, the zona pellucida of oocytes was removed with acid Tyrode’s solution as previously described50,51.
Sperm motility
Cauda epidydimal sperm motility was measured as previously described6,15,48. In brief, sperm were collected from mice and placed in warm human tubal fluid medium in a 37 °C incubator with 5% CO2. A drop of sperm was removed from the suspension and pipetted into a sperm counting glass chamber, then assayed by CASA or video acquisition. CASA was conducted using an IVOS II instrument (Hamilton Thorne) with the following settings: 100 frames acquired at 60 Hz; minimal contrast, 50; 4-pixel minimal cell size; minimal static contrast, 5; 0% straightness (STR) threshold; 10 μm s–1 VAP cutoff; prog. min VAP, 20 μm s–1; 10 μm s–1 VSL cutoff; 5-pixel cell size; cell intensity, 90; static head size, 0.30–2.69; static head intensity, 0.10–1.75; static elongation, 10–94; slow cells motile, yes; ×0.68 magnification; LED illumination intensity, 3,000; IDENT illumination intensity, 3,603; 37 °C. The raw data files (that is, .dbt files for motile sperm and .dbx files for static sperm) were used for sperm motility analyses. For motile sperm, only those for which movement was captured with ≥45 consecutive frames were analysed. For progressive or hyperactivated motility analyses, .dbt files of motile sperm were used as input for CASAnova, as previously described52.
Transmission electron microscopy
Mouse testis and caudal epididymides were dissected and immediately fixed by immersion in Karnovsky’s fixative (2% formaldehyde (v/v) and 3% glutaraldehyde (v/v) in 0.1 M sodium phosphate buffer, pH 7.4; Electron Microscopy Sciences) overnight at 4 °C and washed 3 times in 0.1 M phosphate buffer. Following the third wash, the tissues were post-fixed in 1% osmium tetroxide (w/v; Electron Microscopy Sciences) for 1 h at room temperature, washed 3 more times with water for 10 min each and dehydrated using a graded series of 30%, 50%, 70%, 85%, 95% and 100% (three changes) ethanol and 100% propylene oxide (two changes) and a mixture of 50% propylene oxide (v/v) and 50% SPI-Pon 812 resin mixture (v/v; SPI Supplies). The sample was incubated in seven successive changes of SPI-Pon 812 resin over 3 days, polymerized at 68 °C in flat moulds and reoriented to allow cross-sectioning of spermatozoa in the lumen of epididymis. Sections measuring 70 nm were cut on a Leica EM UC7 ultramicrotome (Leica Microsystems) using a diamond knife, collected on copper mesh grids and stained with 3% lead citrate (w/v) and 0.1% uranyl acetate (w/v) to increase contrast. Finally, sections were examined using a Philips CM10 transmission electron microscope (Philips Electron Optics) at 100 kV. Images were recorded using an Erlangshen digital camera system (Gatan).
piRNA loading and recombinant piRISC purification for MIWI
Recombinant MIWI loading was done as previously described6,15,48. In brief, synthetic piRNA guides (Extended Data Fig. 3) were ordered from IDT and purified by electrophoresis through a 15% denaturing polyacrylamide gel. HEK293T cells (American Type Culture Collection) expressing SNAP-tagged, 3×Flag-tagged MIWI were generated as previously described27. Cells were collected at around 70% confluency using a TC Cell Scraper (ThermoFisher, 50809263) into ice-cold PBS and collected by centrifugation at 500g. Supernatant was removed, and the pellet was stored at −80 °C until lysed in 10 ml of 30 mM HEPES-KOH, pH 7.5, 100 mM potassium acetate, 3.5 mM magnesium acetate, 2 mM DTT, 0.1% (v/v) Triton X-100, 15% (v/v) glycerol and 1× protease inhibitor cocktail (1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma; A8456), 0.3 μM aprotinin, 40 μM betanin hydrochloride, 10 μM E-64 (Sigma; E3132) and 10 μM leupeptin hemisulfate) per gram of frozen cells. Cell lysis was monitored by staining with trypan blue. Crude cytoplasmic lysate was clarified at 20,000g, flash-frozen in liquid nitrogen and stored at −80 °C.
To capture MIWI, 1 ml of clarified lysate was incubated with 20 μl anti-Flag M2 paramagnetic beads (Sigma, M8823) for 4 h or overnight rotating at 4 °C. Beads were washed 4 times with extract buffer (30 mM HEPES-KOH, pH 7.5, 3.5 mM magnesium acetate, 2 mM DTT, 15% (v/v) glycerol and 0.01% (v/v) Triton X-100) containing 2 M potassium acetate and 4 times with extract buffer containing 100 mM potassium acetate. To assemble MIWI piRISC, beads were resuspended in extract buffer containing 100 mM potassium acetate and 100 nM synthetic piRNA guide (Fig. 2a) and incubated with rotation for 30 min at 37 °C or room temperature. After 5 washes in 2 M potassium acetate extract buffer and 5 washes in 100 mM potassium acetate extract buffer, MIWI piRISC was eluted from the beads twice with 200 ng µl–1 3×Flag peptide in 100 µl of 100 mM potassium extract buffer with rotation for 1 h at room temperature. The combined 200 µl eluate was flash-frozen in liquid nitrogen and stored at −80 °C.
Recombinant mouse GTSF1 purification
Recombinant mouse GTSF1 was purified as previously described6,15,48. In brief, pCold-GST(glutathione S-transferase) GTSF-expression vectors were transformed into Rosetta-Gami 2 competent cells (Sigma, 71351). Cells were grown to an OD600 of about 0.6–0.8 in the presence of 1 μM ZnSO4 at 37 °C, then chilled on ice for 30 min to initiate cold shock. Protein expression was induced with 0.5 mM IPTG for 18 h at 15 °C. Cells were collected by centrifugation, washed twice with PBS, and cell pellets were flash frozen and stored at −80 °C. Cell pellets were resuspended in lysis/GST column buffer containing 20 mM Tris-HCl pH 7.5, 500 mM NaCl, 1 mM DTT, 5% (v/v) glycerol and 1× protease inhibitor cocktail (1 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma; A8456), 0.3 μM aprotinin, 40 μM betanin hydrochloride, 10 μM E-64 (Sigma; E3132) and 10 μM leupeptin hemisulfate). Cells were lysed by a single pass at 18,000 psi through a high-pressure microfluidizer (Microfluidics M110P), and the resulting lysate clarified at 30,000g for 1 h at 4 °C. Clarified lysate was filtered through a 0.22 µm Millex Durapore low-protein-binding syringe filter (EMD Millipore) and applied to glutathione Sepharose 4b resin (Cytiva, 17075604) equilibrated with GST column buffer. After draining the flow through, the resin was washed with 50 column volumes GST column buffer. To elute the bound protein and cleave the GST tag in a single step, 50 U HRV3C protease (Millipore, 71493) in 2.5 ml 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM DTT and 5% (v/v) glycerol was added to the column, and the column sealed and incubated for 3 h at 4 °C. Next, the column was drained to collect the cleaved protein. The eluate was diluted to 50 mM NaCl and further purified using a HiTrap Q (Cytiva, 29051325) anion-exchange column equilibrated with 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM DTT and 5% (v/v) glycerol. The bound protein was eluted using a 100–500 mM NaCl gradient in the same buffer. Peak fractions were analysed for purity by SDS–PAGE and the purest were pooled and dialysed into storage buffer containing 30 mM HEPES-KOH, pH 7.5, 100 mM potassium acetate, 3.5 mM magnesium acetate, 1 mM DTT and 20% (v/v) glycerol. Aliquots of the pooled fractions were flash-frozen in liquid nitrogen and stored at −80 °C.
In vitro cleavage assays
Cleavage assays were conducted as previously described6,15,48. In brief, target RNA substrates (Extended Data Fig. 3) were ordered from IDT and labelled using [γ-32P]ATP (Perkin Elmer) and polynucleotide kinase (NEB, M0201). Unincorporated [γ-32P]ATP was removed using a G-25 spin column (Cytiva, 27532501), and target RNA was purified using a 15% denaturing polyacrylamide gel, eluted overnight with rotation in 0.4 M NaCl at 4 °C and collected by ethanol precipitation. Radiolabelled target (0.1 nM final concentration (f.c.)) was added to a mix of purified piRISC (0.5 nM f.c.) and GTSF1 (100 nM f.c.) to assemble a 30 μl cleavage reaction. At 0, 1, 5, 10, 30 and 60 min (0.5, 1, 2, 4 and 6 h for Cox7a2l), a 5 μl sample was quenched in 280 μl 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 25 mM EDTA and 1% (w/v) SDS, then proteinase K (1 mg ml–1 f.c.) was added and the mix incubated at 45 °C for 15 min, followed by extraction with phenol–chloroform–isoamyl alcohol (25:24:1, pH 6.7) and ethanol precipitation. RNA was resuspended in 10 μl 95% (v/v) formamide, 5 mM EDTA, 0.025% (w/v) bromophenol blue and 0.025% (w/v) xylene cyanol, heated at 95 °C for 2 min and resolved in a 7% denaturing polyacrylamide gel. Gels were dried, exposed to a storage phosphor screen and imaged on a Typhoon FLA 7000 (GE). The raw image file was used to quantify the substrate and product bands, corrected for background.
Data were used to fit the burst-and-steady-state scheme
$$E+S\underset{{k}_{-1}}{\overset{{k}_{1}}{\rightleftharpoons }}ES\mathop{\to }\limits^{{k}_{2}}EP\mathop{\to }\limits^{{k}_{3}}E+P,\mathrm{using}\,\mathrm{the}\,\mathrm{equation}:$$
$$[{P}_{{\rm{relative}}}]=f({\rm{t}})=[{E}_{{\rm{relative}}}]({[{k}_{2}/({k}_{2}+{k}_{3})]}^{2}\times (1-{e}^{-[{k}_{2}+{k}_{3}]{\rm{t}}})+[{k}_{2}{k}_{3}/({k}_{2}+{k}_{3})]{\rm{t}})([{E}_{{\rm{relative}}}]).$$
FACS isolation and immunostaining of mouse germ cells
Mouse germ cells were sorted as previously described6,15,48. In brief, testes of 2–7-month-old mice were isolated, decapsulated and incubated for 15 min at 33 °C in 1× Gey′s balanced salt solution (GBSS, Sigma, G9779) containing 0.4 mg ml–1 collagenase type 4 (Worthington, LS004188) rotating at 150 rpm. Seminiferous tubules were then washed twice with 1× GBSS and incubated for 15 min at 33 °C in 1× GBSS with 0.5 mg ml–1 trypsin and 1 µg ml–1 DNase I, rotating at 150 rpm. Next, tubules were homogenized by pipetting through a glass Pasteur pipette for 3 min at 4 °C. Fetal bovine serum (FBS; 7.5% f.c., v/v) was added to inactivate trypsin, and the cell suspension was then strained through a pre-wetted 70 µm cell strainer (ThermoFisher, 22363548). Cells were collected by centrifugation at 300g for 10 min. The supernatant was removed, cells were resuspended in 1× GBSS containing 5% (v/v) FBS, 1 µg ml–1 DNase I and 5 μg ml–1 Hoechst 33342 (ThermoFisher, 62249) and rotated at 150 rpm for 45 min at 33 °C. Propidium iodide (0.2 μg ml–1, f.c.; ThermoFisher, P3566) was added, and cells were strained through a pre-wetted 40 µm cell strainer (ThermoFisher, 22363547). Spermatogonia, primary spermatocytes, secondary spermatocytes and round spermatids were purified using a BD FACSDiscover S8 Cell Sorter (Genomics Core at NYU Center for Genomics and Systems Biology) and a FACSAria II Cell Sorter (BD Biosciences; UMass Medical School FACS Core) as previously described48,53. In brief, the 355-nm laser was used to excite Hoechst 33342, whereas the 488-nm laser was used to record forward and side scatter and to excite propidium iodide. Propidium iodide emission was detected using a 610/20 bandpass filter. Hoechst 33342 emission was recorded using 450/50 and 670/50 band pass filters (Supplementary Fig. 8). Cells were collected by centrifugation at 900g for 10 min. The supernatant was removed and the cell pellets were flash-frozen in liquid nitrogen and stored at −80 °C.
Germ cell stages in the unsorted population and the purity of sorted fractions were assessed by immunostaining aliquots of cells. Cells were incubated for 20 min in 25 mM sucrose and then fixed on a slide with 1% (w/v) PFA containing 0.15% (v/v) Triton X−100 for 2 h at room temperature in a humidifying chamber. Slides were washed sequentially for 10 min as follows: (1) PBS containing 0.4% (v/v) Photo-Flo 200 (Kodak, 1464510); (2) PBS containing 0.1% (v/v) Triton X-100; and (3) PBS containing 0.3% (w/v) BSA, 1% (v/v) donkey serum (Sigma, D9663) and 0.05% (v/v) Triton X-100. After washing, slides were incubated with primary antibodies in PBS containing 3% (w/v) BSA, 10% (v/v) donkey serum and 0.5% (v/v) Triton X-100 overnight at room temperature in a humidified chamber. Rabbit polyclonal anti-SYCP3 (Abcam, ab15093, RRID:AB_301639, 1:1,000 dilution) and mouse monoclonal anti-γH2AX (Millipore, 05-636, RRID:AB_309864, 1:1,000 dilution) were used as primary antibodies. Slides were washed again as described and then incubated with secondary donkey anti-mouse IgG (H+L) Alexa Fluor 594 (ThermoFisher, A-21203, RRID:AB_2535789, 1:2,000 dilution) or donkey anti-rabbit IgG (H+L) Alexa Fluor 488 (ThermoFisher, A-21206, RRID:AB_2535792, 1:2,000 dilution) for 1 h at room temperature in a humidified chamber. After incubation, slides were washed 3 times (10 min each) in PBS containing 0.4% (v/v) Photo-Flo 200 and once for 10 min in 0.4% (v/v) Photo-Flo 200. Finally, slides were dried and mounted in ProLong Gold Antifade mountant with DAPI (ThermoFisher, P36931). To assess the purity of sorted fractions, 50–100 cells were staged by DNA, γH2AX and SYCP3 staining53. All samples used here met the following criteria: spermatogonia, around 95–100% pure with ≤5% pre-leptotene spermatocytes; primary spermatocytes, about 10–15% leptotene/zygotene spermatocytes, around 45–50% pachytene spermatocytes and about 35–40% diplotene spermatocytes; secondary spermatocytes, around 100%; round spermatids, about 95–100%, with ≤5% elongated spermatids.
Small RNA-seq library preparation
Total RNA from sorted mouse germ cells was extracted using a mirVana miRNA isolation kit (ThermoFisher, AM1560). Small RNA libraries were constructed as previously described6,15,48 with modifications. In brief, before library preparation, an equimolar mix of nine synthetic spike-in RNA oligonucleotides (Supplementary Table 9) was added to each RNA sample to enable absolute quantification of small RNAs (Supplementary Table 10). The median volume of primary spermatocytes (1,800 µm3) from ref. 16 was used to calculate the intracellular concentration: 1 molecule per primary spermatocyte corresponds to around 1 pM. To reduce ligation bias and to eliminate PCR duplicates, the 3′ and 5′ adaptors both contained nine random nucleotides at their 5′ and 3′ ends, respectively54 (Supplementary Table 9) and 3′ adaptor ligation reactions contained 25% (w/v) PEG-8000 (f.c.). In brief, 500–1,000 ng total RNA was first ligated to 25 pmol of 3′ DNA adapter (Supplementary Table 9) with adenylated 5′ and dideoxycytosine-blocked 3′ ends in 30 µl of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT and 25% (w/v) PEG-8000 (NEB) with 600 U of homemade T4 Rnl2tr K227Q at 16 °C overnight. After ethanol precipitation, the 50–90 nucleotide (14–54 nucleotide small RNA + 36 nucleotide 3′ UMI adapter) 3′ ligated product was purified from a 15% denaturing urea–polyacrylamide gel (National Diagnostics). After overnight elution in 0.4 M NaCl followed by ethanol precipitation, the 3′ ligated product was denatured in 14 µl water at 90 °C for 60 s, 1 µl of 50 µM RT primer (Supplementary Table 9) was added and annealed at 65 °C for 5 min to suppress the formation of 5′-adapter–3′-adapter dimers during the next step. The resulting mix was then ligated to a mixed pool of equimolar amount of two 5′ RNA adapters (to increase nucleotide diversity at the 5′ end of the sequencing read; Supplementary Table 9) in 20 µl of 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM DTT and 1 mM ATP with 20 U of T4 RNA ligase (ThermoFisher, EL0021) at 25 °C for 2 h. The ligated product was precipitated with ethanol, cDNA synthesis was performed in 20 µl at 42 °C for 1 h using AMV reverse transcriptase (NEB, M0277), and 5 µl of the RT reaction was amplified in 25 µl using AccuPrime Pfx DNA polymerase (ThermoFisher, 12344024; 95 °C for 2 min, 15 cycles of 95 °C for 15 s, 65 °C for 30 s and 68 °C for 15 s; primers are listed in Supplementary Table 9). Finally, the PCR product was purified in a 2% agarose gel. Small RNA-seq libraries samples were sequenced using a NextSeq 550 (Illumina) to obtain 79-nucleotide, single-end reads.
RNA-seq library preparation
Total RNA from sorted germ cells was extracted using a mirVana miRNA isolation kit (ThermoFisher, AM1560). RNA-seq of rRNA-depleted total RNAs was performed as previously described6,15,48,55 with modifications, including the addition of the ERCC spike-in mix to enable absolute quantification of RNAs and the use of unique molecular identifiers in adapters (Supplementary Table 9) to eliminate PCR duplicates54. In brief, before library preparation, 1 µl of 1:100 diluted ERCC spike-in mix 1 (ThermoFisher, 4456740) was added to 1 µg total RNA. To remove rRNA, 1 µg total RNA was hybridized in 10 µl to a pool of 186 rRNA antisense oligos (0.05 µM f.c. each) in 10 mM Tris-HCl (pH 7.4), 20 mM NaCl by heating the mixture to 95 °C, cooling at −0.1 °C s–1 to 22 °C, and incubating at 22 °C for 5 min. RNase H (10 U; Lucigen, H39500) was added and the mixture incubated at 45 °C for 30 min in 20 µl containing 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 20 mM MgCl2. The reaction volume was adjusted to 50 µl with 1× TURBO DNase buffer (ThermoFisher, AM2238) and then incubated with 4 U TURBO DNase (ThermoFisher, AM2238) for 20 min at 37 °C. Next, RNA was purified using RNA Clean & Concentrator-5 (Zymo Research, R1016) to retain ≥200 nucleotide RNAs, followed by the stranded, dUTP-based RNA-seq protocol described in ref. 55. RNA-seq libraries were sequenced using a NextSeq 550 (Illumina) to obtain 79 + 79-nucleotide, paired-end reads. The median number of all non-rRNA transcripts was around 3,400,000 in primary spermatocytes, about 1,700,000 in secondary spermatocytes, about 770,000 in round spermatids and around 50,000 in elongating spermatids. The median volume of primary spermatocytes (1,800 µm3) from a previous study16 was used to calculate intracellular concentration: 1 molecule per primary spermatocyte corresponds to around 1 pM.
For sequencing of polyadenylated RNAs, NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, E7490S) was used to purify poly(A)+ transcripts from 1–2 µg total RNA according to manufacturer’s instructions. Poly(A)+ RNAs were used to prepare RNA-seq libraries with NEBNext UltraExpress RNA Library Prep Kit (E3330S) except that UMI-containing adaptors (Supplementary Table 9) were used. RNA-seq libraries were sequenced using an AVITI benchtop sequencer (Element Biosciences) to obtain 150 + 150-nucleotide, paired-end reads.
Sequencing of 5′ monophosphorylated long RNAs
Total RNA from FACS-purified primary spermatocytes was extracted using mirVana miRNA isolation kit (ThermoFisher, AM1560) and used to prepare a library of 5′ monophosphorylated long RNAs as previously described6,15,16,48,56 with modifications. Briefly, rRNA was depleted as described above for RNA-seq libraries. RNA was ligated to a mixed pool of equimolar amount of two 5′ RNA adapters (to increase nucleotide diversity at the 5′ end of the sequencing read, Supplementary Table 9) in 20 µl of 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM DTT, 1 mM ATP with 60 U of High Concentration T4 RNA ligase (NEB, M0437M) at 16 °C overnight. The ligated product was isolated using RNA Clean & Concentrator-5 (Zymo Research, R1016) to retain ≥200 nucleotide RNAs and reverse transcribed in 25 µl with 50 pmol RT primer (Supplementary Table 9) using SuperScript III (ThermoFisher, 18080093). After purification with 50 µl Ampure XP beads (Beckman Coulter, A63880), cDNA was PCR amplified using NEBNext High-Fidelity (NEB, M0541; 98 °C for 30 s; 4 cycles of: 98 °C for 10 s, 59 °C for 30 s, 72 °C for 12 s; 6 cycles of: 98 °C for 10 s, 68 °C for 10 s, 72 °C for 12 s; 72 °C for 3 min; primers listed in Supplementary Table 9). PCR products between 200–400 bp were isolated from a 1% agarose gel, purified with QIAquick Gel Extraction Kit (Qiagen, 28706), and amplified again with NEBNext High-Fidelity (NEB, M0541; 98 °C for 30 s; 3 cycles of: 98 °C for 10 s, 68 °C for 30 s, 72 °C for 14 s; 6 cycles of: 98 °C for 10 s, 72 °C for 14 s; 72 °C for 3 min; primers listed in Supplementary Table 9). The PCR product was purified from a 1% agarose gel and sequenced using a NextSeq 550 or NovaSeq (Illumina) to obtain 79 + 79-nucleotide or 150 + 150-nucleotide, paired-end reads.
Sequencing of ribosome footprints
Ribosome footprint profiling was performed as described previously57. All steps were performed on ice, unless otherwise indicated. FACS-purified primary spermatocytes, secondary spermatocytes, or round spermatids (1–2 million cells) were lysed in 0.5 ml of 10 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 1% (v/v) Triton X-100, 100 µg ml–1 cycloheximide (Sigma, C4859), and 1× protease inhibitor cocktail (1 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride [Sigma; A8456], 0.3 μM Aprotinin, 40 μM betanin hydrochloride, 10 μM E-64 (Sigma; E3132), 10 μM leupeptin hemisulfate). Cell debris were removed by centrifugation at 20,000g for 10 min at 4 °C. RNase I (Ambion, AM2294) was added to the supernatant (0.2 U μl–1 f.c.) and the sample was incubated at 25 °C for 30 min and then moved to a polycarbonate ultracentrifuge tube (Beckman Coulter, 362305). A 3 ml sucrose cushion (10 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 100 µg ml–1 cycloheximide (Sigma, C4859), 20 U ml–1 SUPERaseIn RNase Inhibitor (Fisher Scientific, AM2694) in 1 M sucrose) was placed under the sample using a 21 G needle (BD, 305167) on a 5 ml syringe (Fisher Scientific, 14955458). Ribosomes were precipitated by centrifugation at about 400,000g for 90 min at 4 °C (100,000 rpm in TLA-110 rotor in Optima MAX-XP Benchtop Ultracentrifuge). RNA was extracted from the ribosome pellet using mirVana miRNA isolation kit (ThermoFisher, AM1560). After ethanol precipitation, the 27–33-nucleotide ribosome footprints were purified from a 15% denaturing urea-polyacrylamide gel (National Diagnostics). After overnight elution in 0.4 M NaCl followed by ethanol precipitation, the 3′ ends of ribosome footprints we dephosphorylated at 37 °C for 4 h in 50 μl of 100 mM MES-NaOH (pH 5.5), 300 mM NaCl, 10 mM MgCl2, 1 U μl–1 SUPERaseIn RNase Inhibitor (Fisher Scientific, AM2694), 15 mM 2-mercaptoethanol, 0.8 U μl–1 T4 PNK (NEB, M0201). After ethanol precipitation, ribosome footprints were ligated to 25 pmol of 3′ DNA adapter for small RNA sequencing (Supplementary Table 9) with adenylated 5′ and dideoxycytosine-blocked 3′ ends in 30 µl of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, and 25% (w/v) PEG-8000 (NEB) with 600 U of homemade T4 Rnl2tr K227Q at 16 °C overnight. After ethanol precipitation, the 5′ ends of 63–69 nucleotide 3′ ligated product (27–33-nucleotide footprints plus 36-nucleotide 3′ unique molecular identifier (UMI) adapter) were phosphorylated in 20 µl of 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT, 1 mM ATP with 20 U of T4 PNK (NEB, M0201). Following an ethanol precipitation, RNAs were denatured in 14 µl water at 90 °C for 60 s, 1 µl of 50 µM RT primer (Supplementary Table 9) was added and annealed at 65 °C for 5 min to suppress the formation of 5′-adapter:3′-adapter dimers during the next step. The resulting mix was then ligated to a mixed pool of equimolar amount of two 5′ small RNA-seq adapters (to increase nucleotide diversity at the 5′ end of the sequencing read, Supplementary Table 9) in 20 µl of 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM DTT, 1 mM ATP with 20 U of T4 RNA ligase (ThermoFisher, EL0021) at 25 °C for 2 h. The ligated product was precipitated with ethanol, cDNA synthesis was performed in 20 µl at 42 °C for 1 h using AMV reverse transcriptase (NEB, M0277), and 5 µl of the RT reaction was amplified in 25 µl using AccuPrime Pfx DNA polymerase (ThermoFisher, 12344024; 95 °C for 2 min, 16 cycles of: 95 °C for 15 s, 65 °C for 30 s, 68 °C for 15 s; primers listed in Supplementary Table 9). Finally, the PCR product was purified in a 2% agarose gel. Ribosome footprint libraries were sequenced using a NextSeq 550 (Illumina) to obtain 79-nucleotide, single-end reads.
GRO-seq
All steps were performed on ice, unless otherwise indicated. FACS-purified primary spermatocytes (1–3 million cells) were collected by centrifugation at 400g for 10 min at 4 °C. Supernatant was removed and cells were carefully resuspended by pipetting in 1 ml swelling buffer (10 mM Tris-HCl (pH 7.5), 3 mM CaCl2, 2 mM MgCl2). Additional 9 ml of swelling buffer was added, then the cells were mixed by swirling and incubated on ice for 5 min. After collecting swollen cells by centrifugation at 400g for 10 min at 4 °C, supernatant was removed and cells were resuspended in 500 μl of Lysis buffer: 10 mM Tris-HCl (pH 7.5), 3 mM CaCl2, 2 mM MgCl2, 10% glycerol, 0.04 U µl–1 RNasin PLUS (Promega, N2615), and 1× protease inhibitor cocktail (1 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma; A8456), 0.3 μM Aprotinin, 40 μM betanin hydrochloride, 10 μM E-64 (Sigma; E3132), 10 μM leupeptin hemisulfate). While carefully swirling the tube, 500 µl of Lysis buffer containing 1% Igepal CA-630 was added by drop and cells were lysed for 5 min on ice. Additional 9 ml of lysis buffer containing 0.5% Igepal CA-630 was added, lysate was mixed by swirling, nuclei were collected by centrifugation at 600g for 5 min at 4 °C, supernatant was removed, and nuclei were resuspended by pipetting in 1 ml of Lysis buffer containing 0.5% Igepal CA-630. Additional 9 ml of Lysis buffer containing 0.5% Igepal CA-630 was added, nuclei were mixed by swirling and collected by centrifugation at 600 × g for 5 min at 4 °C, supernatant was removed, and nuclei were resuspended in 1 ml of Freezing buffer (50 mM Tris-HCl pH 8.0, 5 mM MgCl2, 5 mM EDTA, 40% glycerol, 0.4 U µl–1 RNasin PLUS (Promega, N2615), 1× protease inhibitor cocktail). Nuclei were collected by centrifugation at 900g for 5 min at 4 °C, supernatant was removed, and nuclei were resuspended in 100 μl of Freezing buffer, flash frozen in liquid nitrogen, and stored at −80 °C.
For nuclear run-on reaction, 100 μl of frozen nuclei was thawed on ice for 5 min and then mixed with 100 μl of 10 mM Tris-HCl pH 8.0, 300 mM KCl, 5 mM MgCl2, 10 mM DTT, 0.5 mM of each ATP, GTP, CTP, and BrdUTP (Sigma, B7166), 1% N-Lauroylsarcosine (Sigma L7414), 1 U µl–1 RNasin PLUS (Promega, N2615), and 1× protease inhibitor cocktail. Reaction was mixed with P200 tip with its end cut off and incubated at 30 °C for 30 min, then 24 µl of 10× TURBO DNase buffer and 10 µl of TURBO DNase (2 U µl–1, Fisher Scientific, AM2238) were added, reaction was incubated at 37 °C for 20 min and RNA was extracted with Trizol, resuspended in 30 µl water and stored at −80 °C.
To capture BrdU-labelled nascent transcripts, 30 µl of the sample from previous step was incubated at 65 °C for 5 min, chilled on ice, and mixed with 270 µl of IP buffer: 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.05% Tween-20, 1 U µl–1 RNasin PLUS (Promega, N2615), and 1× protease inhibitor cocktail. Anti-BrdU mouse biotin-conjugated antibody (1 µg, 5 µl of 0.2 µg µl–1 of Clone PRB-1, MilliporeSigma, MAB3262BMI) was added to the RNAs in IP buffer and RNAs were incubated at 4 °C for 1 h with rotation. In a separate tube, 50 µl of Dynabeads MyOne Streptavidin T1 (Fisher Scientific, 6560) were washed at room temperature for 5 min in 1 ml of IP buffer, and beads were then blocked at room temperature for 1 h with rotation in 300 µl of IP buffer containing 0.1% polyvinylpyrrolidone and 1 mg ml–1 Ultrapure BSA (Fisher Scientific, AM2618). After blocking, supernatant was removed, and beads were resuspended in solution containing RNAs and antibody from previous step. Biotin-conjugated antibody was allowed to bind streptavidin beads at 4 °C for 30 min with rotation. Beads were then washed five times with IP buffer at 4 °C for 5 min with rotation, and RNAs were extracted with Trizol. GRO-seq libraries were constructed using the method described for rRNA-depleted RNA-seq libraries and sequenced with a NextSeq 550 (Illumina) to obtain 79 + 79-nucleotide, paired-end reads.
Analysis of small RNA sequencing data
Smal RNA data were analysed as previously described6,15,48. Briefly, the 3′ adapter (5′-TGGAATTCTCGGGTGCCAAGG-3′) was removed with fastx toolkit (v.0.0.14), PCR duplicates were eliminated as described54, and rRNA matching reads were removed with bowtie (parameter -v 1; v.1.0.0; ref. 58) against Mus musculus set in SILVA rRNA database59. Deduplicated and filtered data were analysed with Tailor (v.1.1; ref. 60) to account for non-templated tailing of small RNAs. Sequences of synthetic spike-in oligonucleotides (Supplementary Table 9) were identified allowing no mismatches with bowtie (parameter -v 0; v1.0.0; ref. 58), and the absolute abundance of small RNAs calculated (Supplementary Table 10). Because piRNA 3′ trimming by PNLDC1 results in heterogeneous 3′ ends, sequencing reads were next grouped by their 5′, 25-nucleotide prefix. For further analyses, we kept only prefix groups that met two criteria. First, the prefix group total abundance was ≥1 ppm, that is, ≥10 piRNAs per mouse primary spermatocyte. Assuming a Poisson or a Negative Binomial distribution for piRNA concentration in different cells, this threshold ensures that ≥99.99% of primary spermatocytes contained at least one molecule of the piRNA 25 nucleotide prefix. Second, total abundance of the prefix group was required to be ≥1 ppm in all 12 replicates of the C57BL/6 control samples (Supplementary Table 2). piRNAs were considered undetectable in pi6−/−, pi9−/−, pi17−/−, pi9−/−pi17−/− or pi2−/−pi9−/−pi17−/− primary spermatocytes if their mean abundance in mutants was ≤0.1 ppm.
Analysis of RNA-seq data
RNA-seq data were analysed as previously described6,15,48. Briefly, analysis was performed using piPipes for genomic alignment (v.1.5.0; ref. 61). Briefly, before starting piPipes, sequences were reformatted to extract UMIs54. The reformatted reads were then aligned to rRNA using bowtie2 (v.2.2.0)62. Unaligned reads were mapped to mouse genome mm10 using STAR (v.2.3.1)63, and PCR were duplicates removed54. Transcript abundance was calculated with StringTie (v1.3.4)64 using mm10/rmsk and gene annotation from Ensembl. Differential expression analysis was performed using DESeq2 (v.1.18.1)65. To exclude Cas9-induced off-target changes, only significant (FDR < 0.01) gene expression changes observed in both alleles (em1 and em2) for mice with a pi6−/−, pi9−/− or pi17−/− single mutation and for mice with the pi9−/−pi17−/− mutation were considered (Supplementary Table 3). Thus, all changes in transcript abundance reported for pi9−/−, pi17−/− and pi9−/−pi17−/− mice are the absolute minimums of the two alleles. This approach was not possible for pi2−/− and pi2−/−pi9−/−pi17−/− mice because only one allele of pi2−/− was generated. We considered only transcripts for which abundance was ≥3 TPM (around 10 molecules per primary spermatocytes16), which ensured that, assuming a Poisson or a negative binomial distribution for transcript concentration in different cells, ≥99.99% of primary spermatocytes contained at least 1 molecule of transcript.
Analysis of 5′-monophosphorylated long RNA-seq data
The 5′-monophosphate RNA-seq data were analysed as previously described6,15,48. In brief, data for 5′-monophosphorylated long RNAs was aligned to the mouse genome with piPipes61. In brief, before starting piPipes, the degenerate portion of the 5′ adapter sequences were removed the (nucleotides 1–15 of read1). Because each library was sequenced at least twice to increase the sequencing depth, to harmonize the length of paired-end reads from different runs, sequences were trimmed to 64 nucleotides (read1) + 79 nucleotide (read2) paired reads. The trimmed reads were then aligned to rRNA using bowtie2 (v.2.2.0)62. Unaligned reads were mapped to mouse genome mm10 using STAR (v.2.3.1)63, alignments with soft clipping of ends were removed with SAMtools (v.1.0.0)66, and reads with the same 5′ end were merged to represent a single 5′-monophosphorylated RNA species. For further analyses, only unambiguously mapping 5′-monophosphorylated RNA species were used. For 5′-monophosphorylated RNAs mapped in annotated transcripts, the nucleotide sequence of the corresponding transcript was used to find piRNAs potentially explaining the cleavage, and we used the genomic sequence for 5′-monophosphorylated RNAs mapped outside any annotated transcript. Searches for putative cleavage targets of pi9 and pi17 piRNAs in pi9−/−pi17−/− primary spermatocytes (Fig. 3 and Supplementary Table 6) were performed with a threshold of ≥0.1 ppm for 5′-monophosphorylated putative cleavage products and the following piRNA–target pairing patterns were considered:
-
for piRNAs at ≥1 ppm (10 molecules per primary spermatocyte), ≥20 nucleotides paired between g2 and g25;
-
for piRNAs at ≥5 ppm (50 molecules per primary spermatocyte), contiguous pairing between g3 and g15;
-
for piRNAs at ≥10 ppm (100 molecules per primary spermatocyte), contiguous pairing between g3 and g16;
-
and for piRNAs at ≥50 ppm (500 molecules per primary spermatocyte), contiguous pairing between g4 and g17.
Analysis of ribosome footprint sequencing data
The 3′ adapter (5′-TGGAATTCTCGGGTGCCAAGG-3′) was removed with fastx toolkit (v.0.0.14), PCR duplicates were eliminated as previously described54 and rRNA matching reads were removed with bowtie (parameter -v 1; v.1.0.0)58 against Mus musculus set in the SILVA rRNA database59. Unaligned reads were mapped to mouse genome mm10 using STAR (v.2.3.1)63. Ribosome occupancy was calculated using StringTie (v.1.3.4)64. Differential expression analysis was performed using DESeq2 (v.1.18.1)65. Data from two biological replicates for each pi9em1/em1pi17em1/em1 and pi9em2/em2pi17em2/em2 were obtained and compared against the data from four biological replicates of C57BL/6 controls. Only significant (FDR < 0.01) changes in gene expression observed in both alleles for each pachytene piRNA mutation were considered (Supplementary Table 7). Identification of mRNAs with ELAVL1-binding motif was as previously described22. Data are presented in MS Excel 2013.
Analysis of GRO-seq data
GRO-seq analysis was performed using piPipes for genomic alignment61. In brief, before starting piPipes, sequences were reformatted to extract UMIs54. The reformatted reads were then aligned to rRNA using bowtie2 (v.2.2.0)62. Unaligned reads were mapped to mouse genome mm10 using STAR (v.2.3.1)63 and PCR duplicates were removed54. RNA PolII density was calculated using BEDTools genomecov (v.2.3.4)67,68,69,70,71 as read coverage normalized by sequencing depth and gene length (parts per million per kb; ref. 64). To minimize any contribution from paused RNA PolII, the first 500 bp of genes were excluded from analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.