Cell culture
The human U2OS osteosarcoma cell line (ATCC, HTB-96), HeLa Kyoto cervical carcinoma cell line (CVCL_1922), primary immortalized retinal epithelial cell line hTERT-RPE1 (ATCC, CRL-4000), primary immortalized foreskin fibroblast BJ cells (ATCC, CRL-2522) and their derivatives were grown in Dulbecco’s modified Eagle’s medium (DMEM, high glucose, Glutamax) containing 10% fetal bovine serum (FBS) and penicillin–streptomycin antibiotics (Thermo Fisher Scientific). The E14-derived mouse ES cell lines were obtained from P.-A. Defossez. The J1 mouse ES cell line and PAF15KRKR mutants were obtained from S. Bultmann and H. Leonhardt. Mouse ES cells were cultivated in ESC medium (DMEM (Gibco), 15% KnockOut Serum Replacement (Gibco), 1 mM sodium pyruvate (Gibco), 1× non-essential amino acids (Gibco) and 100 U ml−1 penicillin–streptomycin, 2 mM l-glutamine (Life Technologies), supplemented with LIF (mouse leukaemia inhibitory factor, 10 ng ml−1; Miltenyi) and 0.1 mM β-mercaptoethanol (Sigma)). All cell lines and their derivatives were cultured under standard cell culture conditions (37 °C with 5% CO2, humidified atmosphere). Cells were routinely tested for mycoplasma contamination (MycoAlert, Lonza) and were always negative.
Chemical reagents
Reagents were as follows: hydroxyurea (ribonucleotide reductase (RNR) inhibitor; Sigma-Aldrich, H8627; solubilized in H2O), ceralasertib (AZD6738) (ATR inhibitor; Selleckchem, S7693), adavosertib (MK-1775) (WEE1 inhibitor; Selleckchem, s1525), palbociclib (PD-0332991) (CDK4/6 inhibitor; Selleckchem, S1116), MG132 (proteasome inhibitor; Selleckchem, s2619), bortezomib (proteasome inhibitor; Selleckchem, S1013), PDD 00017273 (PARG inhibitor; Tocris, 5952), doxycycline (Thermo Fisher Scientific, BP2653-5), T2AA (PCNA inhibitor; Tocris, 4723), nocodozole (Tocris, 1228), 5-chloro-2′-deoxyuridine (CldU; Sigma-Aldrich, c6891), 5-iodo-2′-deoxyuridine (IdU; Sigma-Aldrich, I7125), olaparib (PARP inhibitor; Selleckchem S1060), decitabine (DNMT1i; Tocris, 2624), PHA-767491 hydrochloride (CDC7 inhibitor; Tocris, 3140), 5-Ph-IAA (Merck, SML3574) and 5-ethynyl-2′-deoxyuridine (EdU; Thermo Fisher Scientific, A10044). The drugs were reconstituted in DMSO and were used as indicated in the figure legends.
Generation of knockout and complementation cell lines
Knockout of the PAF15 gene in U2OS, HeLa Kyoto and hTERT-RPE1 cells was done using a single guide RNA (gRNA) (targeting exon 2, GGCTGCTCGAGCCCCCAGAA) cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene plasmid 62988, a gift from F. Zhang) via the BbsI restriction site, followed by transfection with Lipofectamine LTX Plus. After two days, transfected cells were selected in medium containing 1–8 µg ml−1 puromycin (InvivoGen, ant-pr-1). After 30–40 h, transfected cells were recovered in plain medium and serially diluted into single cells per well of a 96-well plate to obtain single colonies, expanded and tested for knockout efficiency by immunofluorescence of PAF15 using high-content imaging (QIBC) screening, western blotting and Sanger sequencing of gRNA targeting sites. Only cell lines that passed all validation steps were used. Clones with successful knockout of PAF15 were selected for phenotypic validation. A similar approach was used, by transfecting p53 (also known as TP53) single gRNA (PX459-TP53-exon4, Addgene plasmid 217455, a gift from J. Diffley) or in combination with PX459-PAF15-exon2 (this paper) to generate p53 knockout or p53 + PAF15 double knockout in hTERT-RPE1 cells.
Constitutive and T-REx-inducible cell lines
For complementation assays with constitutive expression, PAF15-KO U2OS cell lines were transfected with the following variant plasmids: PAF15wt-1×MYC-1×-Flag-tag (Origene, RC200694), PAF15 PIP-box mutant (F68F69 to AA mutation)-1×Flag or PAF15 KEN-box mutant (K78A mutation)-1×Flag. Appropriate DNA constructs were transfected using Lipofectamine LTX Plus reagent in PAF15-KO cells. Transfected cells were serially diluted into single cells per well of a 96-well plate to obtain single colonies under selection with DMEM containing geneticin (Gibco, 10131-027) for 12 days. Individual colonies were expanded and tested by immunofluorescence, using QIBC for C terminus Flag-tag or MYC-tag PAF15 (cellular localization), and expression level was tested by western blotting, using antibodies against PAF15.
All of the inducible cell lines were generated through tetracycline-regulated expression of the gene of interest, using T-REx (Thermo Fisher Scientific, K102002). U2OS PAF15-KO cell lines were introduced with doxycycline inducible of each PAF15 wt-1×Flag, PAF15 PIP-box mutant-1×Flag, PAF15 KEN-box mutant-1×Flag, and ΔPAF15 variants (2–11 amino acid deletion) were cloned into the doxycycline-inducible expression vector pcDNA4/TO. PAF15 variant constructs cloned in pcDNA4/TO were co-transfected with the pcDNA6/TR plasmid (expression vector for Tet repressor) by Lipofectamine LTX Plus reagent (Thermo Fisher Scientific, 15338-100). After two days, transfected cells were selected in DMEM supplemented with 10% FBS containing blasticidine and zeocine (Thermo Fisher Scientific; blasticidine, A1113902; zeocine, R25001). After reaching 60–70% confluency, cells were serially diluted into single cells per well of a 96-well plate to obtain single colonies, expanded and tested for PAF15 induction after doxycycline treatment by QIBC, western blotting and high-resolution microscopy.
PAF15 degron cell line
A derivative of the U2OS cell line expressing C-terminally endogenously AID–GFP-tagged PAF15 was generated using CRISPR–Cas9 as previously described59. In brief, gRNA targeting the C terminus of the PAF15 locus (guide: TAACGTCTCCTTGTTTACCC) was cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid 42230, a gift from F. Zhang) via the BbsI restriction site. Cells were co-transfected by Lipofectamine LTX Plus reagent (Thermo Fisher Scientific, 15338-100) with pX330 plasmid containing cloned gRNA, a donor plasmid containing the tag (AID–GFP) with flexible linker flanked by 900-bp homology arms complementary to the C terminus of PAF15 and pCMV6-A-Puro-Tir1-9xMYC plasmid conferring puromycin resistance. After 24 h of transfection, the cells were selected with DMEM containing puromycin (1 µg ml−1) for three days and then serially diluted onto 100-mm dishes. The cells were grown in DMEM to obtain single colonies, which were expanded for further characterization by junction PCR spanning the C terminus of PAF15. Selected clones were functionally validated by immunofluorescence (subcellular localization) using QIBC. Only cell lines that passed all validation steps were used.
PCNA chromobody, TurboID–PCNA and E2F reporter cell lines
U2OS naive cells and PAF15-KO U2OS cells were transfected with plasmid (RFP-pCellCycleChromobody) containing RFP–PCNA chromobody (ChromoTek, ccr) encoding a single-chain antibody to endogenous PCNA. Single clones were selected by geneticin as described in the previous section. Using the same procedure, pBABE-NLS-HA-TurboID-PCNA (Addgene plasmid 215074, a gift from M. Pagano) and pLV-hCDC6p-Venus-puro (Addgene plasmid 212666, a gift from T. Meyer and Y. Konagaya) were introduced into naive U2OS cells. Single clones were selected using puromycin as described above.
Gene silencing by siRNA
Transfections of siRNA duplexes were done with Lipofectamine RNAiMAX (Thermo Fisher Scientific) at a final concentration of 1–20 nM for 48 h (see figure legends for more details). For knockdown of TIMELESS and CLSPN, transfection was performed with 1–5 nM for 24–30 h to prevent adverse cell-cycle effects. The siRNAs were purchased from Thermo Fisher Scientific as Silencer Select reagents targeting the following genes: PAF15 (A: s18863; B: s18862), TIMELESS (s17054), CLSPN (s34330), UHRF1 (A: s26553; B: s26555), LIG1 (s8173), RFC1 (s224528), DNMT1 (s4215) ATAD5 (s36632) or E2F4 (1: 114194; 2: s4415). Non-targeting siRNA from Thermo Fisher Scientific (Ambion negative control 1; 4390844) was used as a control in all experiments.
DNA-fibre analysis
DNA-fibre spreads were performed as described18. In brief, 105 cells were pulse-labelled with 25 µM CldU for the indicated time (see figures for respective labelling protocols), washed three times with DMEM and pulse-labelled with 250 μM IdU with or without the indicated treatment for the indicated time. Labelled cells were collected on ice-cold phosphate-buffered saline (PBS) and mixed with unlabelled cells (1:3). Subsequently, 2 μl of the cell suspension was placed on SuperFrost slides (AB00008032E01MNZ20) and mixed with 8 µl of lysis buffer (0.5% SDS, 200 mM Tris pH 7.5 and 50 mM EDTA) followed by vigorous pipetting for in situ lysis. After 2 min of incubation, slides were tilted to allow the lysate to flow along the slide slowly to the end of the slide. Next, slides were fixed in methanol:acetic acid (3:1) for 12–15 min, washed four times in PBS and transferred to 2.5 M HCl for DNA denaturation for 80 min. Afterwards, slides were neutralized by washing four times in PBS and blocked in blocking buffer (1× PBS, 0.1% Triton X and 1% bovine serum albumin (BSA)) for 5 min. CldU was stained by incubating slides with rat anti-BrdU antibody (Abcam ab6326, 1:100 in blocking buffer) for 90 min. Afterwards, slides were washed once with PBS containing 0.1% Tween, followed by three wash steps with PBS, fixed with 4 % formaldehyde for 12 min and incubated with AlexaFluor 594-conjugated goat anti-rat IgG (1:100; Thermo Fisher Scientific, A-11077) for 60 min. Slides were washed four times with PBS, and IdU was stained using mouse anti-BrdU antibody (1:100; Becton Dickinson, 347580) overnight at 4 °C, followed by AlexaFluor 488-conjugated goat anti-mouse IgG (1:100; Thermo Fisher Scientific; A11029) for 90 min. Fibres were acquired using an Olympus BX53 upright fluorescence microscope with a 40× air objective. For quantifying replication structures, at least 200 DNA fibres were counted per experiment. The lengths of red (CldU) or green (IdU) labelled patches were measured using Fiji ImageJ (National Institutes of Health). Fork speed (in kb min−1) was calculated by multiplying the measured length in μm with a conversion factor of 2.59 kb μm−1 and dividing by the duration of the labelling pulse.
DNA-fibre experiments involving the ssDNA-specific endonuclease S1 were performed as described previously39. In brief, after sequential pulse-labelling with CldU and IdU, cells were collected in ice-cold PBS. Labelled cells were then mixed 1:1 with unlabelled cells and resuspended in hypotonic buffer (10 mM HEPES, pH 7.5, 10 mM NaCl, 0.3 M sucrose and 0.5% Triton X-100) for 15 min on ice, followed by centrifugation at 1,500g for 5 min. Cell pellets were incubated in 50 µl S1 nuclease buffer (30 mM sodium acetate, pH 4.6, 10 mM zinc acetate, 5% glycerol and 50 mM NaCl) with or without 10 U ml−1 S1 nuclease (Invitrogen, 18001-016) for 10 min at 37 °C. After digestion, the cell suspension was resuspended in 50 µl lysis buffer (0.5% SDS, 200 mM Tris, pH 7.5 and 50 mM EDTA) and processed according to the standard DNA-fibre spreading protocol.
To assess the distance between adjacent active replication forks, DNA fibres were prepared as described previously59,60. In brief, CldU-labelled cells were mixed at a 1:10 ratio with unlabelled cells before DNA-fibre preparation. ssDNA was visualized using rabbit anti-ssDNA antibodies (Tecan, IBL International, 18731, 1:500). The distance between active forks was determined by measuring the centre-to-centre spacing between two neighbouring CldU-labelled tracks (referred to as the ‘distance between adjacent active forks’). Local fork density was calculated by counting the number of CldU-labelled tracks within ssDNA-positive DNA fibres and normalizing this value to the length of DNA analysed (Mb).
Immunofluorescence
Cells were grown on round, 12-mm diameter, 1.5-mm-thick glass coverslips (cleaned in 96% ethanol, dried and autoclaved). Unless stated chromatin-bound, cells were washed with ice-cold PBS and fixed in 4% buffered formaldehyde for 12 min at room temperature before permeabilization with PBS containing 0.2% Triton X-100 for 5 min. For assessing chromatin-bound proteins, cells were first pre-extracted with ice-cold PBS containing 0.2% Triton X-100 for 2 min on ice before fixation in 4% buffered formaldehyde for 10 min at room temperature. When Click-iT EdU staining was performed, cells were incubated for 30 min in 10 μM EdU before fixation or pre-extraction. EdU staining was performed according to the manufacturer’s instructions (Thermo Fisher Scientific) before incubating with primary antibodies. All antibodies were diluted in DMEM (high glucose, Glutamax) containing 10% FBS. Primary antibody incubations were performed at room temperature for one hour. Coverslips were washed three times with PBS containing 0.2% Tween (Sigma-Aldrich). Secondary-antibody incubations were performed at room temperature for 30 min and were supplemented with 0.5 mg ml−1 DAPI (Sigma-Aldrich, D8417) to counterstain DNA.
For accessing unligated OkFs, the relevant cells (see also figure legends) were incubated with 10 μM CldU for 48 h to label the parental template DNA. After washing, the cells were exposed to 10 μM EdU for the last 60 min to label nascent DNA—both within the replisome and in post-replication regions—to mark S-phase cells. The cells were then fixed and permeabilized as detailed above and incubated with an alkaline buffer (1×: 50 mM NaOH and 1 mM EDTA in PBS) for 30 min before proceeding with EdU Click-iT staining, immunostaining of CldU (using a rat anti-BrdU antibody, Abcam ab6326, 1:500) and counterstaining of DNA with DAPI as described above.
After three washes in PBS, coverslips were washed twice in distilled water, dried on 3-mm paper and mounted in 4.5 ml Mowiol-based mounting medium (containing Mowiol 488 (Calbiochem), glycerol and Tris-HCl, pH 8.5). For all of the confocal and STED imaging, slides were mounted with Prolong Diamond Antifade Mountant (Thermo Fisher Scientific, P36961).
QIBC
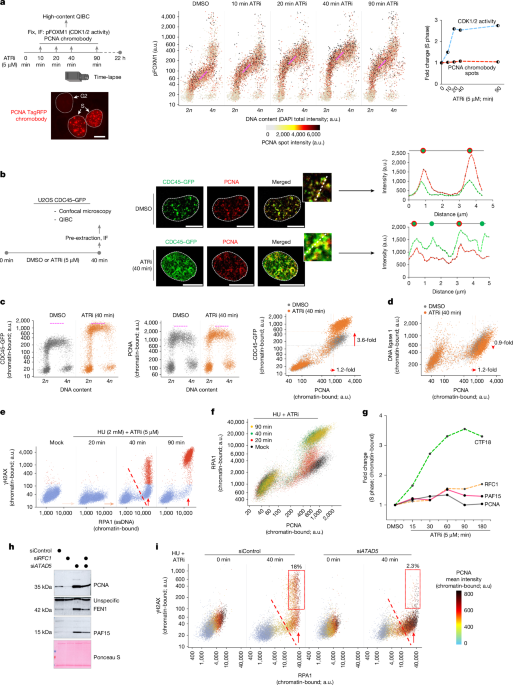
QIBC was performed as previously described2,18,39,59,61. In brief, images were acquired with a ScanR inverted microscope high-content screening station (Olympus) equipped with wide-field optics, a 203, 0.75-NA (UPLSAPO 203) air objective, fast excitation and emission filter-wheel devices for DAPI, FITC, Cy3 and Cy5 wavelengths, an MT20 illumination system and a digital monochrome Hamamatsu ORCA-R2 CCD camera (yielding a spatial resolution of 320 nm per pixel at 203 and binning of 1). Images were acquired in an automated manner with the ScanR acquisition software (Olympus, 3.4). Depending on cell confluency, 100 images were acquired, containing more than 5,000 cells per condition. Acquisition times for the different channels were adjusted for nonsaturated conditions in a 12-bit dynamic range, and identical settings were applied to all the samples in one experiment. Images were processed and analysed with ScanR analysis software. First, a dynamic background correction was applied to all images. The DAPI signal was then used to generate an intensity-threshold-based mask to identify individual nuclei as main objects. This mask was then applied to analyse pixel intensities in different channels for each nucleus. After segmentation of nuclei, foci were segmented as above, and the desired parameters for the different nuclei or foci were quantified, with single parameters (mean and total intensities, foci count and foci intensities) as well as calculated parameters (sum of foci intensity per nucleus). These values were then exported and analysed with TIBCO v.12.4. This software was used to quantify absolute, median and average values in cell populations and to generate all colour-coded scatter plots. Within one experiment, similar cell numbers were used for the different conditions (at least 2,000–10,000 cells), and for visualization, low x-axis jittering was applied (random displacement of objects along the x axis) to make overlapping markers visible.
Confocal microscopy
Confocal images were acquired with a Nikon A1 confocal Ti2 microscope integrated with a 100 × 1.45 NA oil, Plan Apochromat λ objective and NIS-Elements AR software (v.5.20.02). A resonant scanner equipped with an A1-DUG hybrid 4-channel detector was used for image acquisition at 512 × 512 pixels. Laser power, detector gain and exposure time were appropriately adjusted with identical settings applied within a series of experiments. Microscope performance and channel alignment were regularly checked by imaging of 200-nm multicolour fluorescent beads.
STED
For combined STED and confocal microscopy, EdU-labelled pre-extracted samples were imaged using an Abberior Facility Line STED microscope (equipped with a 100× NA 1.4 oil objective (UPLSAPO100X, Olympus). Click-iT EdU coupled to AlexaFluor 488 was imaged for confocal microscopy but PAF15 (using STAR RED goat anti-rabbit (1:250, STRED-1007, Abberior) was imaged for both confocal and STED microscopy. Regions of interest (5× 5 µm2) showing immobilized dye signal were imaged for the indicated number of frames using standard confocal and STED imaging conditions, 10 µs dwell time, 50 nm pixel size and a repetition frequency of 40 MHz. To collect emission spectra, 488-nm and 561-nm laser lines were used with a power of 10 µW as measured at the sample plane. All data were acquired using the iMSPECTOR v.16.3.13787 acquisition software. A mean filter with a radius of one pixel was applied to remove background noise.
SCAR-seq
PAF15 SCAR-seq was performed as described33 with the following modifications. Cells were grown to 80% confluence and EdU was added for 20 min (final concentration 20 µM) to label newly synthesized DNA and cross-linked by adding formaldehyde to a final concentration of 1% for 10 min, then the reaction was quenched with glycine (final concentration 0.1 M) and washed twice with ice-cold PBS. Fixed cells were lysed in ice-cold lysis buffer at room temperature for 20 min. Lysis buffer was prepared by adding one-third dilution buffer (100 mM Tris-HCl pH 8.6, 100 mM NaCl, 5 mM EDTA and 5.0% Triton X-100) and two-thirds SDS buffer (100 mM NaCl, 50 mM Tris-HCl pH 8.1, 5 mM EDTA and 0.5% SDS) supplemented with cOmplete protease inhibitor (Roche, 11873580001). Chromatin extracts were sonicated using Diagenode Bioruptor Pico to generate chromatin fragments averaging 250–300 bp. For each chromatin immunoprecipitation (ChIP), approximately 300 µg of chromatin, based on DNA concentration, and 4 µg of anti-PAF15 antibody (Santa Cruz sc390515; PCLAF) were used. The PAF15–DNA complexes were pulled down using anti-mouse IgG Dynabeads (Thermo Fisher Scientific, 11201D), and the DNA chromatin complexes were de-cross-linked at 55 °C for four hours. One per cent of the lysate was reserved as an input control.
Click biotinylation of newly replicated DNA, new-strand isolation and libraries were prepared with xGEN UDI-UMI adapters (IDT). Libraries were sequenced in paired-end mode on an Illumina NextSeq 2000.
SCAR-seq data processing
Raw reads were trimmed. Reads with poor quality (lower than 20) were filtered using cutadapt (v.2.6) and aligned to the mouse reference genome (GRCm39) using bowtie2 (v.2.4), and duplicated reads were marked and removed by Picard tools (v.3.4).
The resulting processed bam files were split into forward and reverse strands according to SAM flags (SAMtools, v.1.13). Forward strands were defined with both -f 83 and -f 163 SAM flag, and reverse strands with -f 99 and -f 147. SAM flags were inverted according to the inverted orientation of IDT UDI-UMI adapters. Read coverage was computed using multiBamSummary (deepTools, v.3.5.4) in bins of 1 kb. Bins with normalized read counts per million (CPM) < 0.3 were excluded from the following analyses.
PAF15 strand-specific partitioning was computed from mapped binarized files according to the following: Partitioning = (F − R)/(R + F), where F and R correspond to the number of mapped reads to the forward strand and to the reverse strand, respectively.
OK-seq
OK-seq was performed as described previously34, with modified adapter sequences as follows:
Adapter mix A:
Adapter1w-mixA: [SpC3] ACACTCTTTCCCTACACGACGCTCTTCCGATCT
Adapter1c-mixA: [SpC3] NNNNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT
Adapter2w-mixA: [Phos]AGATCGGAAGAGCACACGTCTGAACTCCAGTCA [SpC3]
Adapter2c-mixA: [SpC3] TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNN [SpC3]
Libraries were sequenced in paired-end mode on an Illumina NextSeq 2000.
OK-seq data processing
Raw reads were trimmed. Reads with poor quality (lower than 20) were filtered using cutadapt (v.2.6) and aligned to the mouse reference genome (mm39) using bowtie2 (v. 2.4), and duplicated reads were marked and removed by Picard tools (v.3.4). The resulting processed bam files were split into forward and reverse strand according to SAM flags (SAMtools, v.1.13). Forward strands were defined using both -f 99 and -f 147 SAM flags, and reverse strands with -f 83 and -f 163. Read coverage was computed using multiBamSummary (deepTools, v.3.5.4) in bins of 1 kb. Bins with normalized CPM < 0.3 were excluded from the following analyses. The RFD score was computed from a binarized file according to the following: RFD = (R − F)/(R + F), where R and F correspond to the number of forward and reverse mapped reads, respectively. Initiation zone positions were computed with only the regions with RFDmax > 0 and RFDmin < 0 were considered. Initiation zones with a minimum size of 10 kb, an efficiency higher than 10% (ΔRFD > 0,2; ∆RFD is the difference between RFDmax and RFDmin) and a minimum overlap of 50% between 2 biological replicates were retained, resulting in 4,559 regions.
SCAR-seq and OK-seq data analysis
Average profiles of PAF15 partition and RFD values around initiation zones were computed up to 100 kb upstream and downstream of each initiation zone by averaging values within each bin position and smoothed by considering neighbouring windows of 15 kb. The difference in PAF15 partitioning at the left and right ends of the initiation zone was tested with a paired Wilcoxon signed-rank test in three independent biological replicates.
Genome-wide Spearman’s rank correlation was calculated in 1-kb bins and represented as a hexplot. For IGV visualization, PAF15 strand partitioning values and OK-seq RFD values were smoothed considering the neighbouring 15 bins.
FRAP
U2OS cells expressing GFP-tagged-PCNA were seeded in imaging dishes (Nunc, Lab-Tek, 155361) and before imaging, transferred to CO2-independent medium. FRAP data were acquired using a Nikon A1 Ti2 microscope with a 60× 1.2 NA Plan Apo Water Immersion objective and NIS-Elements (v.5.30.02) software, under stable temperature conditions of 37 °C. After ten pre-bleaching frames (pre), a single bleach pulse (488-nm argon laser set to 100% power) was delivered in a defined region, followed by time-lapse imaging for 3 min at maximum scanning speed (six frames per second) with the laser transmission attenuated to 2.5%. Image analysis was performed by first extracting the mean GFP-associated fluorescence intensity for each time point in the following regions: bleaching region (Ifrap(t)); background outside the nucleus (Iback(t)); and signal within the nucleus in which bleaching was performed (Iref(t)). After background correction, double normalization was applied, which corrects for differences in the starting intensity in the Ifrap region and for loss in total nuclear fluorescence in the Iref region owing to the bleaching pulse and to acquisition bleaching.
Cell synchronization
U2OS cells were synchronized at the G2/M phase by the addition of nocodazole. Exponentially growing U2OS cells were incubated with 200 ng ml−1 nocodazole for 20 h. For enrichment of cells into different cell-cycle phases, cells were washed and cultured in fresh DMEM. The cells were collected at 0, 6, 8, 10, 12, 15 and 24 h and fixed with 4% buffered formaldehyde for 10 min at room temperature and permeabilized with PBS containing 0.2 % Triton X-100 for 5 min. QIBC-based PAF15 total pool and cell-cycle analysis was performed with Click-iT EdU staining, assessing sequential cell-cycle phase transition. Complementary to this, a western blot was performed to assess the PAF15 in soluble and chromatin-bound subcellular fractions.
RNA isolation for RNA-seq and quantitative PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen), following the manufacturer’s protocol. RNA concentration was measured using a NanoDrop spectrophotometer. cDNA was prepared, according to the manufacturer’s instructions, using the AMPIGENE cDNA Synthesis Kit (Enzo Life Sciences). The quantitative PCR (qPCR) reactions were performed in triplicate using iTaq Universal SYBR Green Supermix (Bio-Rad). Relative expression levels were calculated using the 2−ΔΔCT method.
The following primer pairs were used:
ATAD5 forward (5′-GCCAACCCTTCGAAACATCTG-3′) and reverse (5′-CTTCAAAATAGTGCAGGAATCTTCT-3′), GAPDH forward (5′-CACCATCTTCCAGGAGCGAG-3′) and reverse (5′-TGATGACCCTTTTGGCTCCC-3′), PCNA forward (5′-GCAGATGTACCCCTTGTTGT-3′) and reverse (5′-ATCCTCGATCTTGGGAGCCA-3′) and PAF15 forward (5′-GGCGGGATAGTTTTCGGGTC-3′) and reverse (5′-CGAGCAGCCACCACTTTTCT-3′).
For DMSO and CDK4/6 inhibitor, three biological repeats were performed in hTERT-RPE1 cells. RNA-seq was performed at BGI Genomics using DNBseq stranded mRNA libraries generated on the DNBseq NGS platform after quality control. The quality of the raw sequencing data was assessed using FastQC (v.0.11.9) and MultiQC (v.1.10.1). The raw bulk RNA-seq data were aligned to the human genome assembly (GCF_000001405.39_GRCh38.p13), and differential gene expression was analysed using the DEF analysis plan and a Poisson distribution model at BGI Genomics.
For control and E2F4 depletion, three biological repeats were performed in hTERT-RPE1 cells. RNA-seq was performed according to the manufacturer’s instructions (TruSeq2, Illumina) using 500 ng of RNA for the preparation of cDNA libraries. Sequencing reads were mapped to the human genome (hg38) using STAR, and tag counts were summarized at the gene level using HOMER56, allowing only one read per position per length. TiCoNE25 was used to cluster differentially expressed genes as determined by DESeq2.
Clonogenic survival assay
Naive cells, gene-knockout cells and cells stably expressing PAF15 variant constructs were transfected with control and other siRNAs for 48 h; these cells were seeded in six-well plates in triplicate (200, 500 and 1,000 cells per well). After 24 h, genotoxic treatments were performed as indicated in the figure legends. Cells were incubated for ten days, fixed with 4% formaldehyde and stained with crystal violet. Individual colonies were counted manually, and the percentage survival was calculated as the value for an indicated siRNA divided by the value for the control siRNA, after correcting for the respective plating efficiency.
PLA
PLA was performed as described previously18 with modifications. In brief, cells were fixed either with methanol (for PCNA interactions) for 15 min or 4% buffered formaldehyde (PAF15 interactions) for 12 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. Cells were then blocked for one hour in DMEM containing 10% FBS and incubated with primary antibody in a humidity chamber for one hour and secondary-antibody probes in a humidity chamber at 37 °C for one hour. In situ proximity polymerization followed by ligation was performed using a Duolink Detection Kit (Sigma-Aldrich), and the nucleus was counterstained with DAPI. Nuclear foci were imaged using a ScanR inverted microscope and processed for QIBC. At least 5,000 cells per condition were analysed in each experiment.
Chromatin fractionation
A total of 3 × 106 cells were collected from experimental conditions as described in the figure legends. The cells were washed with PBS and collected. The soluble protein fraction was removed by incubation in 0.5% Triton X-100 in PBS supplemented with 1× protease and phosphatase inhibitors (PPi) cocktail (Roche). The fractions were centrifuged for 5 min at 4 °C at 16,000g. The samples were washed with PBS containing 0.5× PPi. Finally, the cell pellets were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton X-100 and 0.5% deoxycholate) (Sigma-Aldrich, R0278-500ML), with benzonase (Merck, E1014) and 100 μg ml−1 RNaseA (Thermo Fisher Scientific, EN0531). The samples were sonicated at low amplitude, on ice for two repeats of 20-s pulses after incubation for one hour on ice. Chromatin-bound protein pools were collected by centrifugation at 4 °C, 16,000g for 30 min. Protein concentration was quantified using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, 23227), and 20–50 μg protein from chromatin fraction was used for western blots.
Western blotting
Whole-cell extracts (WCE) were obtained by lysis in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0 % IGEPAL CA-630, 0.1% SDS and 0.1% sodium-deoxycholic acid), supplemented with protease and phosphatase inhibitors (Roche) containing benzonase (Novagen). Protein extracts from WCE or chromatin fractions were separated by SDS–PAGE after boiling samples in reducing buffer (DTT and β-mercaptoethanol) as per standard procedures. Separated proteins were transferred from the gel to a nitrocellulose membrane. The membrane was blocked for one hour in TBS 0.1% Tween containing 5% powdered milk (TBS-T) and subsequently incubated with primary antibodies for two hours at room temperature or overnight at 4 °C. Phospho-specific primary antibodies were diluted in 3% BSA in TBS-T solution. Secondary peroxidase-coupled antibodies (Vector Labs) were incubated at room temperature for 1 h. Enhanced chemiluminescence (ECL)-based chemiluminescence was detected with an Amersham Imager 680 system (software v.2.0).
Immunoprecipitation
A total of 2 × 107 cells from naive U2OS, PAF15 constitutive overexpression and doxycycline-induced PAF15 cells with the indicated siRNAs (see figure legends) were collected for WCE, or the chromatin-bound proteins were either processed without cross-linking or cross-linked by incubating cells in 0.1% formaldehyde for 15 min at room temperature. The reaction was quenched by incubating with 0.125 M glycine. Cells were then collected by scraping and washed with PBS and incubated on ice for 15 min with 0.5% Triton X-100 in PBS supplemented with PPi cocktail (Roche). The samples were washed with PBS containing 0.5× PPi, and the nuclear pellets were resuspended in RIPA lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% SDS, 1% Triton X-100 and 0.5% deoxycholate), with benzonase (Merck, E1014) and 100 µg ml−1 RNaseA (Thermo Fisher Scientific, EN0531). After incubation for one hour on ice, samples were sonicated at low amplitude on ice for two repeats of 20-s pulses. After centrifugation at 16,000g for 30 min, the supernatant was collected, and proteins were quantified using the Pierce BCA Protein Assay kit. Then, 1,000 μg of the chromatin or WCE was incubated overnight at 4 °C with anti-MYC magnetic beads (Sigma) or anti-Flag magnetic agarose (Thermo Fisher Scientific). Similar reactions were also performed for naive U2OS cells with an equivalent amount of beads, and 5–10% of the chromatin was used as an input control. Bound proteins were eluted from beads by boiling in NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, NP0007) with NuPAGE Sample Reducing Agent, or eluted using Flag peptide (Thermo Fisher Scientific). Total immunoprecipitants were then analysed by immunoblotting or processed for MS analysis as specified in the figure legends.
Chromatin-bound PAF15 immunoprecipitation and MS
Chromatin-bound PAF15 immunoprecipitation protein products were resolved on a NuPAGE Novex Bis-Tris 4–12% gel (Invitrogen). Lanes for each sample were sliced (around 1 mm3) and gel slices were destained further with a buffer containing 50 mM ammonium bicarbonate and 50% acetonitrile. Gel pieces were dehydrated by the addition of 100% acetonitrile. The gel pieces were incubated with 10 mM DTT for 30–45 min and further incubated for 30 min with 55 mM IAA and subsequently dehydrated with 100% acetonitrile. The proteins were digested with trypsin (Sigma) at 37 °C for 16 h, after which the samples were acidified with 0.5% trifluoroacetic acid (TFA). The resulting peptides were desalted on reversed-phase C18 StageTips columns, eluted with 40 ml of 50% acetonitrile, 0.1% TFA followed by 10 ml of 70% acetonitrile, 0.1% TFA and subsequently dried by vacuum centrifugation. Samples were redissolved in 0.1% formic acid of which 5 of 12 µl were loaded onto an Easy nLC (Thermo Fisher Scientific) equipped with a custom-made two-column set-up (pre-column: 100 µm ID, 3.5 cm, Reposil-Pur 120 C18-AQ, 5 µm (Dr. Maisch); analytical column: 75 µm, 18 cm, Reposil-Pure 120 C18-AQ, 3 µm (Dr. Maisch)). Peptides were eluted with a gradient of solvent B (95% acetonitrile, 0.1% FA) as specified below, with solvent A being 0.1% formic acid, and sprayed directly into an Exploris 480 (Thermo Fisher Scientific) mass spectrometer. The gradient was constructed as follows: 5% to 25% B in 70 min, 25% to 40% B in 19 min and 40% to 95% B in 1 min.
On the Exploris 480, MS1 data were obtained at resolution 120 K, scan range 350–1,600, AGC target 2.5e10 and with the Max IT set to auto. MS2 data were recorded as top 12 at a resolution of 30 K, AGC target 2e5, Max IT 100 ms and 20-s dynamic exclusion. Data were searched against the SwissProt database of human proteins using Proteome Discoverer 2.5 (Thermo Fisher Scientific) with Mascot 2.7 as the search engine. The m/z tolerance was set to 5 ppm for precursors and 0.05 Da for fragment ions. Quantification was done as label-free quantification based on the area under the curve of the (up to) top five most abundant peptides for each protein, also using match between runs.
Proximity labelling of TurboID–PCNA and MS
Stably expressing PCNA–TurboID U2OS cells treated with control and PAF15 siRNA were grown to 80% confluency, and proximity labelling was done with 50 µM biotin for 30 min. Collected cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate, 1% (v/v) Triton X-10, pH 7.5), sonicated, cleared by centrifugation and incubated with streptavidin magnetic beads (Pierce) in RIPA buffer for enrichment of biotinylated proteins. The beads were sequentially washed with RIPA buffer, Tris buffer (50 mM Tris, 150 mM NaCl, pH 7.5), 1 M KCl, 0.1 M Na2CO3 and 2 M urea in 10 mM Tris pH 7.5, using a KingFisher system (Thermo Fisher Scientific).
After enrichment of proximity labelled proteins or chromatin, proteins were precipitated on streptavidin magnetic beads (Pierce) or HILIC microparticles (Resyn BioSciences), respectively, in 70% acetonitrile, reduced, alkylated and washed sequentially in 95% acetonitrile and 70% ethanol to remove detergents62 and digested on beads with trypsin and Lys-C. The resulting peptides were desalted on StageTips and subjected to LC–MS analysis on an EASY nano-LC 1200 system (80–120-min gradients) coupled to an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific). MS data were acquired by data-dependent acquisition and subjected to MaxQuant database searches63 using a human Uniprot-reviewed sequence database, match between runs and label-free quantification.
AlphaFold-based modelling of protein structures
AlphaFold 3 with its implementation in the AlphaFold Server was used to predict the DNA–protein structures40. PyMOL was used for structure modelling and generating high-resolution images. The protein sequences were extracted from the Uniprot database. The input DNA sequence was extracted from the experimentally determined structure of human PCNA with DNA (PDB: 7QO1)64. All five AlphaFold-predicted models were aligned or superimposed using the PyMOL software and only models that showed consistency throughout the models are present in the manuscript. PCNA or its yeast homologue POL30 was used as a reference for PyMOL alignment or superimposition, and only the models showing a root mean standard deviation (RMSD) lower than 1.0 are used throughout the manuscript. The electrostatic surface visualization of PCNA–DNA–PAF15 was performed with a plug-in installation of APBS in PyMOL65. The interpretation of predicted alignment error plots generated by AlphaFold 3 was performed with the PAE viewer66. The PAE plots were further exported as .svg and .png files and only the latter were annotated in Adobe Illustrator to obtain the final figures.
ABC modelling and analysis of E2F occupancy on PCNA and PAF15 causal enhancers
We downloaded IDR-thresholded peak lists from ENCODE67 and generated a consensus peak list using iterative overlap peak merging68. We counted reads in peaks using UCSC tools and z-transformed the counts. To predict causal enhancers in cell type at the PCNA and PAF15 loci, we used the ABC model69, using the z-transformed chromatin accessibility as a proxy for enhancer activity and a power transformation of the distance between the enhancer and the transcription start site as a proxy for contact frequency, as suggested in the ABC paper. We selected enhancers with an ABC score greater than 0.2 as putative causal enhancers. At these enhancers, we calculated the average occupancy of E2F1 and E2F4 and tested for significant differences between the average occupancy across cell types using a paired t-test. List of ENCODE DCC experiment accession numbers: ChIP–seq: E2F1 in K562 (ENCSR153DWR), ChIP–seq: E2F4 in K562 (ENCSR368GJN), ATAC-seq: K562 (ENCSR868FGK), ChIP–seq: E2F1 in HepG2 (ENCSR717ZZW), ChIP–seq: E2F4 in HepG2 (ENCSR924LSO), ATAC-seq: HepG2 (ENCSR291GJU), ChIP–seq: E2F1 in MCF7 (ENCSR000EWX), ChIP–seq: E2F4 in MCF7 (ENCSR505NMN), ATAC-seq: MCF7 (ENCSR422SUG), ChIP–seq: E2F1 in HeLa-S3 (ENCSR000EVJ), ChIP–seq: E2F4 in HeLa-S3 (ENCSR000EVL) and DNase-seq: HeLa-S3 (ENCSR959ZXU).
Public single-cell RNA-seq data analysis
For investigation of single-cell PAF15 expression in human breast and kidney cancer, features, barcodes and raw read counts from two publicly available datasets were retrieved from the Gene Expression Omnibus repository GSE17607870 and the Human Cell Atlas project Haniffa-Human-10x3pv271. Both datasets were pre-processed using the pipeline suggested by Seurat (v.4.0.3)72. Low-quality cells with fewer than 200 detected features and mitochondrial gene contributions lower than 20% were removed from the kidney single-cell data. Subsequently, normalization, scaling and identification of variable features were done on both datasets using SCTransform73 regressing out mitochondrial, ribosomal and haemoglobin gene percentage. Doublets were estimated and removed using scDblFinder (v.1.2.0)74. Batch integration was performed with Harmony (v.1.2.0)75. Dimensional reduction by uniform manifold approximation and projection (UMAP) was recalculated using the integrated lower-dimensional space, using the first 50 principal components and with nearest neighbours set to 15. The original Louvain algorithm was used for community detection. Automated cell annotation of clusters was done with CellTypist (v.1.6.3)76 using the Adult_Human_Kidney and Cells_Adult_Breast models for kidney and breast tissue cells, respectively.
The Cancer Genome Atlas data analysis
Data were extracted from the The Cancer Genome Atlas (TCGA) TARGET GTEx database. For survival analysis, raw read counts were retrieved from all open-access TCGA projects under the data category transcriptome profiling using TCGAbiolinks (v.2.32.0)77. Raw counts were batch-corrected using ComBatseq from the sva package (v.3.52.0)78. Patients were split on the basis of the best-performing cut-off in gene-expression level between the lower and upper quartiles using Cox proportional hazards regression modelling. Patients were stratified by high and low expression of PAF15 and PCNA. Cut-off values were chosen on the basis of the most significant values from Cox regression on all values from lower to upper quartiles. An estimate of a survival curve was computed using Kaplan–Meier with the survival package (v.3.7.0). For gene-expression analysis in normal and cancer tissue, RSEM-normalized expression data from the TCGA TARGET GTEx cohort (n = 19,109) were retrieved from the UCSC Xena platform79. All statistics on TCGA-derived data were performed in R, and data visualization was done with ggplot2 (v.3.5.1)80.
Reproducibility
For all QIBC, DNA-fibre, proteomics and RNA-seq experiments, a minimum of three technical repeats and a minimum of two biological replicates were performed. Experiments were not randomized, and no blinding was used during data analysis. In box plots, the centre lines are medians, the boxes indicate the 25th and 75th centiles and the whiskers indicate 5% and 95% values. Sample sizes, statistical tests and numbers of replicates for all image-based experiments are specified in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.