Cell culture
Cell lines
A549 (CCL-185, American Type Culture Collection (ATCC)), BSR T7/5 (ref. 55) (BHK-21 cells that constitutively express the T7 RNA polymerase, RRID:CVCL_RW96) and HEK293T cells (CRL-3216, ATCC) were cultured in DMEM (31966021, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; F7524, Merck) and 1% penicillin–streptomycin (pen–strep; 15140122, Thermo Fisher Scientific). Vero cells (CCL-81, ATCC) were cultured in DMEM supplemented with 5% FBS and 1% pen–strep. HEp-2 cells (CCL-23, ATCC) were cultured in MEM (42360032, Thermo Fisher Scientific) with 10% FBS and 1% pen–strep. All cells were maintained at 37 °C and 5% CO2. Cell lines used in this study were confirmed to be mycoplasma negative.
Primary cells
Primary human nasal epithelial cells (HNECs) from healthy donors (EP51AB, Epithelix) were cultured in an air–liquid interface (ALI) transwell system, as previously described56. In brief, HNECs were expanded to log phase in PneumaCult Ex Plus medium (05040, StemCell Technologies). Passage 1 cells were cryopreserved in 78% Ham’s F12 (51651C, Merck/Sigma), 10% heat-inactivated FBS, 2% HEPES (pH 7.2) and 10% DMSO, then stored at −135 °C. After thawing, cells were expanded with medium, replaced every 1–2 days, then seeded at a density of 7 × 104 cells per well on PET transwells (CLS3470, Corning; 0.4-μm pores, 6.5 mm in diameter) with 100 μl apical and 650 μl basolateral PneumaCult Ex Plus medium. After 48–72 h, confluent monolayers were airlifted by aspirating media and adding PneumaCult ALI medium (05001, StemCell Technologies) supplemented with 100 U ml−1 penicillin, 0.1 mg ml−1 streptomycin (P4333, Sigma), 0.48 μg ml−1 hydrocortisone (07926, StemCell Technologies) and 4 μg ml−1 heparin (07980, StemCell Technologies) to only the basolateral chambers. Medium was replaced every 2–3 days; from 2 weeks post-airlift, apical surfaces were washed 1–2 times weekly with 200 μl HBSS (14025050, Gibco) to remove mucus. Cultures were maintained at 37 °C with 5% CO2.
Nasal wash samples
Nasal wash samples from individuals infected with RSV were obtained from participants enrolled in a previously described controlled human infection model (CHIM) study57. That study, conducted independently of the present work, involved inoculation of healthy adults with RSV-A Memphis-37b58. Nasal wash samples were collected at defined time points, frozen and stored for subsequent analysis. For the present study, nasal wash samples collected from a single participant on study days 4 and 5, which showed detectable nasal viral load as determined by quantitative PCR with reverse transcription (RT–qPCR), were utilized.
RSV protein samples
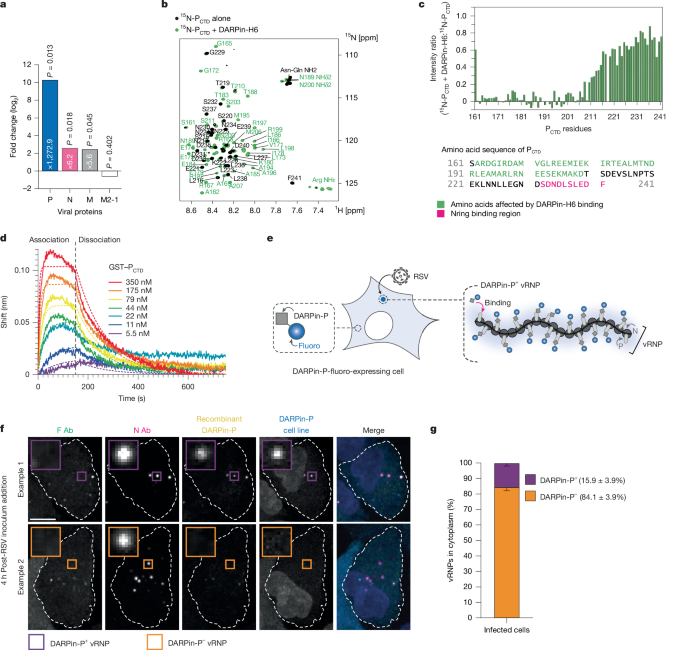
RSV Nrings without tag were purified by co-expression in Escherichia coli and co-purification with GST–PCTD (amino acids 161–241), using the GST tag, as previously described19. When specified, the GST tag was removed by thrombin cleavage. For NMR measurements, 15N-labelled PCTD was expressed in E. coli from PCTD cloned into the pGEX-4T3 plasmid, and purified as previously described59. For expression and purification of all the GST–P fragments, the corresponding P sequences were cloned into pGEX-4T3 plasmid, and the proteins were expressed in E. coli. Purifications were performed as previously described19.
Screening of DARPins binding to RSV vRNP
To generate DARPin binders for the GST–PCTD + Nrings complex, the biotinylated GST–PCTD + Nrings complex was immobilized alternatingly on either MyOne T1 streptavidin-coated beads (65601, Thermo Fisher Scientific) or Sera-Mag neutravidin-coated beads (78152104011150, Cytiva) depending on the selection round. Ribosome display selection was performed as previously described60, but using a semiautomatic KingFisher Flex MTP96 well platform.
The fully synthetic library includes N3C-DARPins with three randomized internal repeats with the original randomization strategy as reported61, but including a stabilized C-cap18,62,63. In addition, the library is a 1:1 mixture of DARPins with randomized and non-randomized N-caps and C-caps, respectively18,64, and successively enriched pools were ligated in a ribosome display-specific vector60. Selection was performed over four rounds with decreasing concentrations of biotinylated GST–PCTD + Nrings complex (250 pmol, 125 pmol and 5 pmol) for the first three rounds and 50 pmol of target for the last recovery round, and increasing washing steps60,65. For rounds 2–4, prepanning with biotinylated GST was used to remove potential binders against GST.
To screen individual DARPins for their binding properties, the selected pool of DARPins from ribosome display was subcloned by restriction digest with BamHI and HindIII into the pQE30-derived bacterial expression vector pQIq (Qiagen). This creates DARPins with an N-terminal MRGS(H6) tag and a C-terminal FLAG-M2 tag. Single DARPin clones (n = 192) were screened against the GST–PCTD + Nrings complex and GST only, both directly immobilized, by a crude extract ELISA. The crude extracts were prepared as previously described66. Thirty-two identified DARPin clones were sequenced and 13 of these clones were unique in their sequence. These 13 clones were IMAC purified and validated in an ELISA, as previously described66, against the GST–PCTD + Nrings complex and GST.
Purified DARPin-P production
DARPin-H6 with an N-terminal MRGS(H6) tag and a C-terminal FLAG-M2 tag (His6–DARPin–H6–FLAG, referred to as DARPin–H6 in Fig. 1 and DARPin-P thereafter, originally obtained as 011-1055-C6-2605-H5 in the selection), was cloned into the pQIq vector backbone67 (Supplementary Table 1). The plasmid was further modified to generate fluorescent protein fusions: DARPin-P–BFP, DARPin-P–sfGFP and DARPin-P–mRuby3 (Supplementary Table 1). DARPin-P and the fluorescent fusion proteins were recombinantly expressed in E. coli XL-1 blue cells (200249, Agilent) and purified using the MRGS(H6) tag. In brief, a 20 ml primary culture of transformed cells was grown overnight in 2× YT medium (Y2377-250G, Merck) supplemented with 1% glucose and 100 µg ml−1 ampicillin (A5354-10ML, Merck) at 37 °C with shaking at 160 rpm. The primary culture was diluted into 400 ml of the same medium and incubated at 37 °C with shaking at 160 rpm until the OD600 reached 0.5–0.8. Protein expression was induced with 0.5 mM IPTG (AM9464, Thermo Fisher Scientific), and the culture was further incubated at 37 °C and 160 rpm for 4 h. Cells were harvested by centrifugation at 4,000g for 20 min and stored at −20 °C until use. The cell pellets were thawed and resuspended in 25 ml of buffer A (PBS, pH 7.2, supplemented with 150 mM NaCl and 30 mM imidazole) containing 5% (v/v) glycerol and cOmplete Mini EDTA-free Protease Inhibitor Cocktail (11836170001, Merck). Cells were lysed by sonication, and the lysate was clarified by centrifugation at 20,000g for 30 min. The resulting clear supernatant was loaded onto an Ni-NTA agarose column (R90115, Thermo Fisher Scientific), which was subsequently washed with 20 column volumes (CV) of buffer A. Bound MRGS(H6)-tagged proteins were eluted using buffer A supplemented with 400 mM imidazole. Fractions containing the recombinant protein were pooled, and the imidazole was removed using a PD SpinTrap G-25 Desalting Column (28918004, Cytiva). Purified proteins were stored as single-use aliquots at −80 °C until further use.
DARPin-P characterization
Mass spectrometry
DARPin-P immunoprecipitation on RSV virions for mass spectrometry
Sucrose purified RSV (4.4–9.2 × 107 plaque-forming units (PFU)) per condition were lysed in ice-cold RIPA lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate and 1% Triton X-100) supplemented with 100 µg ml−1 AEBSF serin protease inhibitor (78431, Thermo Fisher Scientific). Purified DARPin-P (12.5 µg) was adsorbed with 10 µl anti-DYKDDDDK magnetic agarose (A36797, Thermo Fisher Scientific) for 1 h at 4 °C in RIPA lysis buffer under mild rotation. DARPin-P-adsorbed beads or untreated beads were washed in RIPA lysis buffer before incubating with virus lysate for 2 h at 4 °C followed by five washes in ice-cold RIPA lysis buffer. Proteins were eluted in 1% SDS in D-PBS at 50 °C under agitation. For proteomic analysis, three replicates were generated.
Immunoprecipitation for phosphorylation mass spectrometry
A549 cells (7.5 × 105) infected with RSV for 24 h were lysed in ice-cold RIPA lysis buffer supplemented with 100 µg ml−1 AEBSF serin protease inhibitor and 1× PhosSTOP (4906837001, Roche). Purification was performed either with anti-RSV P (polyclonal; see details below) for total P or with DARPin-P as follows: for total P, 10 µg anti-RSV P (polyclonal) antibody was incubated with whole-cell lysate for 1 h at 4 °C in RIPA lysis buffer under mild rotation. Protein A and protein G magnetic beads (50 µl 1:1; 10002D and 10004D, Thermo Fisher Scientific) were washed and added for 1 h at 4 °C, washed three times with ice-cold RIPA and eluted in 1% SDS in D-PBS at 50 °C under agitation. For DARPin-P purification, 12.5 µg purified DARPin-P was adsorbed with 10 µl Pierce anti-DYKDDDDK magnetic agarose (A36797, Thermo Fisher Scientific) for 1 h at 4 °C in RIPA lysis buffer under mild rotation. DARPin-P-adsorbed beads were washed in RIPA lysis buffer before incubating with whole-cell lysate for 2 h at 4 °C followed by three washes in ice-cold RIPA lysis buffer. Proteins were eluted in 1% SDS in D-PBS at 50 °C under agitation. For phospho-proteomic analysis, three replicates were generated.
Protein digestion for proteomics analysis
Immunoprecipitation eluates were precipitated onto 400 µg of carboxyl-coated beads (45152105050250 and 65152105050250, Cytiva), washed and digested according to a standard solid-phase-enhanced sample preparation (SP3) protocol68 using 50 ng of sequencing grade trypsin (V5111, Promega).
Phospho-enrichment
Following digestion, samples were desalted using the Oasis HLB 96-well μElution Plate (186001829, Waters), as per the manufacturer’s instructions. Peptides were eluted in 25 μl of elution (phospho-enrichment loading solvent) containing 80% acetonitrile, 1 M glycolic acid, 6% trifluoroacetic acid (TFA) and 0.4 mg of pre-equilibrated MagReSyn Zr-IMAC beads were added to each sample (MR-ZRM002, Resyn Biosciences). Samples were incubated for 20 min (at room temperature at 1,350 rpm), then washed once in 400 μl of loading solvent, twice in 400 μl wash buffer 1 (30% acetonitrile and 1% TFA), and twice in 400 μl wash buffer 2 (10% acetonitrile and 0.1% TFA). Phospho-peptides were then eluted with 4% ammonium hydroxide for 10 min (at room temperature at 1,350 rpm). The elution step was repeated twice more, and eluates were acidified with formic acid to a final concentration of 5%. The collected peptides were then desalted using the Oasis HLB plate and eluted once in 25 μl SPE elution buffer (50% acetonitrile and 0.1% formic acid in water) and then diluted in 100 μl 5% DMSO and 5% formic acid before liquid chromatography–tandem mass spectrometry analysis.
Mass spectrometry
Analysis of immunoprecipitation peptides was carried out using an Ultimate 3000 nano-LC 1000 system coupled to an Orbitrap Exploris (Thermo Fisher Scientific). Phospho-enriched peptides were analysed using an Ultimate 3000 nano-LC 1000 system coupled to an Orbitrap Ascend Tribrid Mass Spectrometer (Thermo Fisher Scientific). Peptides were initially trapped on a C18 PepMap100 pre-column (300 μm inner diameter × 5 mm, 100 A) and then separated on an in-house built C18 column (Reprosil-Gold, Dr. Maisch, 1.9-μm particle size) column (ID: 50 μm, length of 50 cm) at a flow rate of 100 nl min−1. Total immunoprecipitation peptides were separated over 60 min (10–33% B) and phospho-enriched peptides were separated over 30 min (10–36% B). In both cases, mobile phase A (water, 5% DMSO and 0.1% formic acid) and mobile phase B (acetonitrile, 5% DMSO and 0.1% formic acid) were used. Separated peptides were directly electrosprayed into the mass spectrometer. Mass spectra were acquired in the Orbitrap (350–1,400 m/z, resolution of 60,000, automatic gain control (AGC) target of 3 × 106 and maximum injection time of 50 ms) in a data-dependent mode. For total immunoprecipitation samples, the top 40 most-abundant peaks in the survey scan were fragmented using higher-energy collisional dissociation (HCD, resolution of 7,500, AGC target of 4 × 104 and maximum injection time of 64 ms). For phospho-enriched samples, the top 30 most-abundant peaks in the survey scan were fragmented using HCD (MS2 scans were acquired using the ion trap analyser in Turbo mode; AGC target of 2 × 104 and maximum injection time of 35 ms).
Phosphorylation analysis of DARPin-P-isolated RSV P
A549 cells (5 × 105) infected with RSV for 24 h were lysed in ice-cold RIPA lysis buffer supplemented with 100 µg ml−1 AEBSF serin protease inhibitor and 1× PhosSTOP (4906837001, Roche). Purification was performed either with anti-RSV P (polyclonal) for total P or with DARPin-P as follows: for total P, 10 µg anti-RSV P (polyclonal) antibody was incubated with 50 µl 1:1 protein A and protein G magnetic beads (10002D and 10004D, Thermo Fisher Scientific) for 1 h at 4 °C, washed with D-PBS and crosslinked to the beads with 2.5 mM BS3 (A39266, Thermo Fisher Scientific) for 30 min at room temperature before quenching with 50 mM Tris pH 7.4 for 15 min. After washing three times in RIPA, beads were incubated with whole-cell lysate for 1 h at 4 °C, three times with ice-cold RIPA and eluted in 1% SDS in D-PBS at 50 °C under agitation. For DARPin-P purification, 12.5 µg purified DARPin-P was adsorbed with 10 µl Pierce anti-DYKDDDDK magnetic agarose (A36797, Thermo Fisher Scientific) for 1 h at 4 °C in RIPA lysis buffer under mild rotation. DARPin-P-adsorbed beads were washed in RIPA lysis buffer before incubating with whole-cell lysate for 2 h at 4 °C followed by three washes in ice-cold RIPA lysis buffer. Proteins were eluted in 1% SDS in D-PBS at 50 °C under agitation. Three replicates were generated. For phosphorylation analysis 50 µM or 100 μM Phos-Tag-acrylamide (TA9H9A175FCA, Merck) containing 12% SDS–PAGE, gels were generated, samples separated at 180 V and washed in transfer buffer with 1 mM EDTA followed by wash in transfer buffer before being transferred to nitrocellulose membranes. Samples were blocked in 3% milk in D-PBS-T before probing with anti-RSV P (polyclonal) at a 1:1,000 dilution, using donkey anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody, Alexa Fluor Plus 680 (A32802, Thermo Fisher Scientific) for detection with a Li-COR Odyssey Fc.
NMR
NMR experiments were carried out on a Bruker 800 MHz Avance III spectrometer equipped with a TCI cryoprobe. Protein samples were dialysed into PBS at pH 6.4 and mixed or diluted to obtain the desired final concentration. All samples contained 7.5% D2O as a lock substance. 1H–15N HSQC spectra were acquired on samples with 15N-PCTD (50 µM) at a temperature of 288 K. The GST purification tag was removed from 15N-PCTD, which ensured that the protein was monomeric and precluded any steric hindrance that might interfere with binding. NMR data were processed with TopSpin 4.0 (Bruker) software and analysed with CcpNMR Analysis software69. Amide assignment of 15N-PCTD was previously performed (Biological Magnetic Resonance Bank entry ID 26906)59.
Native agarose gel electrophoresis
Samples in the presence of 50% sucrose loading buffer were loaded on native 1% agarose gel and migration was performed in 1× Tris–glycine buffer during 1 h 30 min at 80 V before staining with amido black 10B70.
BLI
Purified MRGS(H6)-tagged DARPin-P (ligand) was diluted in BLI assay buffer (PBS + 0.01% bovine serum albumin (BSA) + 0.002% Tween 20, pH 7.4) at room temperature. Ligand at 20 µg ml−1 was loaded on His1K (anti-penta-his) biosensors (Sartorius) for 150 s. Kinetic experiments were performed at 30 °C with 1,000 rpm shaking in 96-well black plates using the Octet Red 96e system (Fortebio). Biosensors loaded with DARPin-P were successively equilibrated for 60 s in assay buffer (baseline step), incubated in a dilution of the analyte GST–PCTD (twofold from 350 nM to 5.5 nM) for 150 s (association step), then incubated in assay buffer for 600 s (dissociation step). One ligand-bound sensor was incubated in assay buffer as a reference to measure signal drift. As references for binding specificity, biosensors in the absence of ligand were used in a parallel kinetics experiment. Real-time binding kinetics were analysed and calculated using the Octet Red software package (v9.0). Raw signal was processed using the double-reference method, by subtracting both the biosensors without ligand (unspecific signal) and the signal in the absence of analyte (drift), after baseline alignment and interstep correction at the dissociation. Kinetic modelling was done by analysing association and dissociation signals using global fitting with a 1:1 model.
Reporter cell line generation
Plasmids for lentiviral vectors
Self-inactivating lentiviral vectors were based on the pHR vector backbone under the control of the spleen focus-forming virus (SFFV) promoter. The WPRE element was omitted to aid in obtaining low transgene expression levels (pHR-pSFFV-insert-ΔWPRE). The sequences of the inserted transgenes are listed in Supplementary Table 1.
Lentiviral transduction
All cell lines that stably express transgenes were generated via lentiviral transduction. Unless stated otherwise, transgenes were introduced into A549 cells. Lentivirus was produced by polyethylenimine transfection (23966, Polysciences) of lentiviral plasmid carrying the transgene of interest and the helper plasmids pMD2G and psPAX2. Three days after transfection the supernatant containing lentivirus was collected and filtered (0.45-µm filter) to remove cellular debris, and was added to recipient cells of interest along with 10 mg ml−1 polybrene (sc-134220, Santa Cruz Biotechnology) and subject to spin infection for 120 min at 2,000 rpm at 25 °C. Following spin infection, the medium was refreshed, and after two passages, single cells from polyclonal cell populations were sorted in 96-well plates by FACS. The fluorescence expression level for each individual cell line was carefully determined by initial investigation on the polyclone followed by screening of multiple monoclones. In general, reporter cell lines were selected with low levels of cytoplasmic fluorescence (to have minimal fluorescence background and suppress aggregate formation). Furthermore, we only selected cell lines in which RSV infection kinetics were not affected compared with parental cell lines. If a cell line required the expression of multiple transgenes, lentiviral transduction was either carried out simultaneously (maximally two different lentiviruses) or sequentially (minimally starting from 3 days following spin infection). The specific cell lines used in each experiment is listed in Supplementary Table 1.
RSV strains, design, production and validation
Strains
Unless otherwise stated, human RSV, subgroup A, strain Long, (ATCC VR-26, GenBank AY911262.1; referred to as RSV (Long strain)) was used. In addition, the laboratory strains, human RSV-A2 (ATCC VR-1540) and RSV-98-25147-X (GenBank FJ948820; referred to as RSV-X) and two RSV subgroup A clinical isolates (03-036465 (GenBank JQ901457.1) and 18-0011989) were used for specified experiments. The RSV clinical isolates were provided by M. C. Viveen and F. Coenjaerts, and further information about their isolation, assessment and storage has been previously described71.
Design
All viral sequences were derived from RSV (Long strain). Engineered RSV strains were designed based on a previously established recombinant human RSV reverse genetics system in which unique restrictions sites have been introduced between individual RSV genes to facilitate cloning of the viral genome (reverse genetics vector pACNR-rHRSV)17.
The recombinant RSV virus encoding for an additional mCherry fluorescent protein (located between the P and M genes on the viral genome) encoded by the pACNR-rHRSV-mCherry was previously described17.
The genome expression construct encoding for recombinant upstream and downstream SunTageng RSV, were generated by designing introduction of the SunTag gene inserts at the MluI restriction site between P and M genes and BstEII restriction site between F and M2 for the upstream and downstream strains, respectively. The introduced SunTag genes were designed to contain the same gene-regulatory elements, gene start (GS), gene end (GE) and the 5′ and 3′ untranslated regions (UTRs), as the N gene. Furthermore, the reporter genes were introduced such that the gene-regulatory elements of the upstream and downstream genes with respect to the insert location were not disrupted. The coding sequence of the reporter genes contained a translation start codon in an optimal Kozak sequence (GCCACCATGG), followed by a sequence encoding a SunTag array and downstream gene (to generate a longer transcript allowing for more ribosomes to simultaneously translate the SunTag-encoding mRNAs, which results in higher fluorescence signal of translation sites). The upstream reporter carries a 24×SunTag–BFP and the downstream reporter carries a 12×SunTag–Kif18b. The sequences of the additional SunTag genes introduced are provided in Supplementary Table 1.
The engineered RSV strains carrying endogenously tagged RSV proteins (P and L) were designed by targeting regions in the respective gene sequence that have previously been shown to be conducive for insertions with minimal disruption to viral function. An ALFA tag was inserted into the coding sequence of P at amino acid 72 (ref. 37) to generate a P-ALFA-tageng RSV and an HaloTag was inserted into the coding sequence of L at amino acid 1749 (refs. 72,73) to generate a L-HaloTageng RSV. All insertions were flanked by flexible linker sequences (ALFA tag flanked by 5′ and 3′ six-amino-acid-long linkers and HaloTag flanked by a 5′ 18-amino-acid and 3′ 14-amino-acid linker). The engineered gene sequences are provided in Supplementary Table 1.
Generation of plasmids encoding recombinant RSV strains
The pACNR reverse genetics vectors containing the RSV genomes with the upstream and downstream SunTag genes were generated by Gibson assembly and correct insertion verified by sequencing the insert and the gene upstream and downstream of the insert location.
The reverse genetics vectors carrying the P-ALFA-Tageng and L-HaloTageng RSV strains were generated by transformation-associated recombination (TAR) cloning in yeast as previously described74. In brief, overlapping PCR products containing the RSV genome, ALFA tag/HaloTag and TAR vector (pCC1BAC-His3)75 were transformed into the yeast strain VL6-48N76. Yeast-assembled full-length clones were isolated, amplified in E. coli strain EPI300 and sequences were verified by full-plasmid nanopore sequencing (Plasmidsaurus), where remaining uncertainties in the viral sequence were confirmed by Sanger sequencing.
RSV rescue and concentration
Recombinant WT and tagged RSV strains were rescued by reverse genetics and amplified in HEp-2 cells as previously described77. In brief, the appropriate pACNR reverse genetics vector (1.25 µg) was co-transfected together with the expression vectors pCITE-N (1 µg), pCITE-P (1 µg), pCITE-L (0.5 µg) and pCITE-M2-1 (0.25 µg)17,78, each under the control of the T7 promoter72 in BSR T7/5 using Lipofectamine 3000 (L3000001, Thermo Fisher Scientific), according to the manufacturer’s protocol. Three days following transfection, virus was collected and efficiency of rescue assessed by plaque assay carried out on HEp-2 cells. The viruses were amplified for a subsequent 3–4 passages on HEp-2 cells at an MOI of 0.01 PFU per cell to minimize formation of defective viral genomes. For the P-ALFA-tageng and L-HaloTageng RSV strains, the rescue protocol was similar, with the exception that the helper plasmids were in pcDNA3 (NR-36462, NR-36463, NR-36464 and NR-36461, BEI Resources), transfection of BSR T7/5 cells was performed using Lipofectamine LTX (A12621, Thermo Fisher Scientific), passaging was performed on Vero cells, and viruses were titrated by 50% tissue culture infective dose (TCID50) assay. The final stocks were grown on Hep-2 cells. RSV-A2 and RSV-X were propagated in HEp-2 cells and Vero cells, respectively.
Viral supernatants from passage 3–4 were precleared for cells and debris by centrifugation and subsequently concentrated. Concentration was done either via polyethylene glycol-mediated precipitation (viral supernatant was stirred with 10% PEG6000 (50 mM Tris-HCL, pH 7.4, 150 mM NaCl and 1 mM EDTA) for 3 h at 4 °C, followed by centrifugation at 3,500 RCF for 30 min at 4 °C), or by pelleting through a 10% sucrose-containing buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl and 0.5 mM EDTA; centrifuge viral supernatant in 10% sucrose buffer (4:1 v/v ratio) at 10,000 RCF for 4 h at 4 °C). The virus containing pellets were resuspended in 10% sucrose in PBS, snap-frozen and stored at −80 °C as single-use aliquots. RSV-A2 and RSV-X were purified between layers of 10% and 50% sucrose by ultracentrifugation, washed with plain RPMI and concentrated on 100-kDa filters (UFC910024, Millipore) by centrifugation. The virus was washed from the filters in phenol red-free RPMI (11835-030, Gibco) supplemented with 10% sucrose and subsequently snap-frozen and stored at −80 °C.
Viral growth assays
Viral growth assays were performed to assess the fitness of the engineered RSV strains with respect to their WT counterpart.
The upstream and downstream SunTageng RSV strains growth kinetics were assessed on A549 cells. A549 cells were inoculated with virus at an MOI of 3 PFU per cell (in DMEM + 10% FBS + 1% pen–strep) for 1 h, following which cells were washed in PBS and supplemented with fresh media (DMEM + 10% FBS + 1% pen–strep). New progeny virus production was assessed up to 5 days post-infection by collecting samples at multiple time points along the time course. The viral titres of the samples were determined by plaque assays on HEp-2 cells.
The growth kinetics of the P-ALFA-tageng and L-HaloTageng RSV strains were assessed on Vero cells via qPCR. Cells were infected with an MOI of 0.04 PFU per cell and supernatant was collected up to 48 h post-infection in MagNA Pure External Lysis Buffer (06374913001, Roche) for RNA isolation. Sample (120 μl) was mixed with 50 μl AMPure XP Reagent (A63880, Beckman Coulter), loaded in a 96-well PCR plate, washed three times with 70% ethanol using a DynaMag-96 Side Skirted Magnet and RNA was eluted in 50 μl RNase-free H2O. RNA, 5 μl per 20 μl reaction volume, was reverse transcribed and amplified in one reaction using the TaqMan Fast Virus 1-step Master Mix (4444432, Thermo Fisher Scientific) according to the manufacturer’s protocol on a StepOnePlus System. RSV-A primers with a FAM probe were used (Supplementary Table 1) and a serially diluted RSV-A plasmid was included as a reference.
Assessing RSV P phosphorylation mutations
RSV P phosphorylation mutants, in which the phosphorylation sites overlapping the DARPin-P binding sites were mutated, were generated on the pCITE-P plasmid backbone. The sequences of the inserted phospho-mutant P sequences are listed in Supplementary Table 1. BSR T7/5 cells were transfected with WT P and mutants at 70% confluence in a 96-well glass-bottom plate (MGB096-1-2-LG-L, Brooks Life Science Systems and 89627, Ibidi) with 1 µg of plasmid DNA using Lipofectamine 3000 Transfection Reagent (L3000015, Thermo Fisher Scientific) following the manufacturer’s protocol. Twenty-four hours post-transfection, cells were fixed and subject to downstream analysis.
Assessing RSV pseudo-VFs
BSR T7/5 cells were transfected at 70% confluence in a 96-well glass-bottom plate (MGB096-1-2-LG-L, Brooks Life Science Systems and 89627, Ibidi) with a plasmid mixture containing pCITE-P (1 µg) and pCITE-N (1 µg) using Lipofectamine 3000 Transfection Reagent (L3000015, Thermo Fisher Scientific) following the manufacturer’s protocol. Twenty-four hours post-transfection, cells were fixed and subject to downstream analysis.
RSV infection for imaging-based studies
Inoculum for RSV infection
One day before imaging, the required A549 cell lines were seeded on 96-well glass-bottom plates (MGB096-1-2-LG-L, Brooks Life Science Systems and 89627, Ibidi) such that cells were at approximately 80–90% confluency at the point of viral inoculum addition. For fixed-cell experiments, the viral inoculum was made in cell culture medium (DMEM + 10% FBS + 1% pen–strep). For live-imaging experiments, viral inoculum was made in imaging medium (CO2-independent Leibovitz’s-L-15 media (21083027, Thermo Fisher Scientific) supplemented with 10% FBS and 1% pen–strep).
For live-cell assessment of RSV infection progression and success, a 1:1,000 dilution of RSV glycoprotein G antibody (133; referred to as AbG-fluoro; Janelia Fluor 646, NBP2-50411JF646 and Alexa Fluor 405, NBP2-50411AF405, NOVUS Biologicals) was added directly into the viral inoculum. For visualization of the L-HaloTageng RSV, the viral inoculum was supplemented with 5 nM HaloTag ligand JFX650 (ref. 79). Both fluorescent media supplementations gave negligible fluorescence background and was maintained on cells until experimental end point. The details of viral inoculums used in each experiment is listed in Supplementary Table 1.
Nasal wash samples were added undiluted onto the relevant A549 cell lines (as described in the figure legend and Supplementary Table 1) and maintained for 4 h at 37 °C, following which cells were rinsed in PBS and topped with warmed imaging media before imaging.
For ExM, WT A549 cells were grow on 12-mm coverslips (no. 1.5H) such that cells were at approximately 90% confluence at the point of viral inoculum addition. Cells were infected with WT RSV (MOI of 2) in the presence of the translation inhibitor emetine (see below) in cell culture medium. At 4 hpi, cells were fixed, stained and subject to downstream analysis.
MOI used for imaging-based studies
For imaging studies, MOI was based on the fraction of cells containing vRNPs at 6 h following viral inoculum administration. In fixed-cell experiments with WT A549 cells, this was assessed by the presence of N+/F− foci (antibody staining), and in live-cell experiments, this was assessed in the Pexo-fluoro transgenic cell line by the presence of Pexo foci. The MOI value established using this imaging-based method is three orders of magnitude higher than the MOI calculated using plaque assay-based viral titration on HEp-2 cells (0.1 imaging-based MOI relates to 0.0001 PFU per cell). Unless otherwise specified, the imaging-based MOI used was between 0.05 and 0.25.
Chemical inhibitors
The following inhibitors were used in this study: PC786 (15 µM, RSV polymerase inhibitor; HY-102038, MedChemExpress), TMC353121 (180 nM, RSV fusion inhibitor; HY-11097, MedChemExpress), emetine dihydrochloride (emetine; 50 µg ml−1; inhibits translation by preventing translocation of the tRNA–mRNA complex; 324693, Merck), cyclopamine (CPM; 4 µM; inhibits RSV infection by disorganizing and hardening VFs; HY-17024, MedChemExpress)37,38 and puromycin dihydrochloride (puromycin; 0.1 mg ml−1; a tyrosyl-tRNA mimic that blocks translation by releasing elongating polypeptide chains; A1113803, Thermo Fisher Scientific). When used, PC786 and emetine were added directly with the viral inoculum, and TMC353121, CPM and puromycin were added to infected cells at indicated time points. Following addition, the inhibitors were maintained on cells until experimental end point. For multi-day inhibitor treatment protocols, the inhibitors were refreshed after each 24 h.
Assessing infecting vRNPs
Assessment of the viral protein and RNA levels on infecting RSV vRNPs were carried out in the presence of the translation inhibitor emetine. Emetine containing inoculum was maintained on cells for 1 h, following which the RSV fusion inhibitor TMC353121 was added to the inoculum to prevent subsequent infections. Three hours post-fusion inhibitor addition, the cells were assayed, either by live imaging or following fixation and subject to antibody or smFISH staining protocols. This time point was chosen to ensure that all infecting vRNPs had fully separated such that infecting individual vRNPs could be assessed.
Generation and assessment of RSV passaged in different cell types
HNECs cultured in an ALI transwell system were fully differentiated for 5–7 weeks, with maturity confirmed by ciliary beating and mucous production. Three days before infection, basal medium was replaced with complete PneumaCult ALI medium without heparin and hydrocortisone (ALI infection medium). On the day of infection, apical surfaces were washed once with 200 μl HBSS and incubated for 10 min at 37 °C. The apical wash and basal medium were aspirated, and 650 μl fresh ALI infection medium was added to the basal compartment. RSV (Long strain) was diluted in HBSS, and 200 μl of the inoculum was applied per transwell to achieve 1 × 104 TCID50 per well. Two wells per donor were infected and incubated for 1 h at 37 °C. The inoculum was aspirated, and cells were washed three times with 200 μl HBSS. Cultures were then maintained at 37 °C and 5% CO2. Apical washes were collected at 24, 48 and 72 hpi by adding 200 μl HBSS, incubating for 10 min at 37 °C, and collecting the wash for viral quantification. Washes were mixed with sucrose to a final 10% concentration, aliquoted, snap-frozen and stored at −80 °C.
Vero, HEp-2 and A549 cells were seeded in 48-well plates (in the appropriate media, see above), with two wells per cell line per experimental repeat. RSV inoculum was prepared by diluting RSV (Long strain) in MEM. Culture medium was removed, and 100 μl inoculum was added to achieve 1 × 104 TCID50 per well. Cells were incubated with the inoculum for 3 h at 37 °C, then inoculum was aspirated, and cells were washed twice with PBS. Fresh medium containing 2% FBS was added. At 24, 48 and 72 hpi, supernatants were collected, mixed with sucrose to 10% final concentration, aliquoted, snap-frozen and stored at −80 °C.
The 72 hpi viral supernatant samples were used for subsequent analysis. Viral supernatants were titrated on the Pexo-fluoro A549 cell line and used at an MOI of 0.05.
Immunofluorescence
A detailed step by step protocol for immunofluorescence on different samples can be found in Supplementary Information.
Antibody conjugation
RSV nucleoprotein antibody (RSV3132 (B023); referred to as anti-RSV N; NB100-64752, NOVUS Biologicals) was fluorescently labelled using N-hydroxysuccinimide (NHS)-esters (Alexa Fluor 555 NHS ester and Alexa Fluor 647 NHS ester; AF555, A20009, AF647 and A20106, Thermo Fisher Scientific) for use in immunofluorescence. The antibody was buffer exchanged and concentrated with a 10 kDa MWCO PES concentrator (88535, Thermo Fisher Scientific) to yield 3 mg ml−1 antibody solution in 7.5% sodium bicarbonate buffer, pH 8.3 (25080094, Thermo Fisher Scientific). The dye-NHS stocks were prepared in anhydrous DMSO (AM9342, Thermo Fisher Scientific). A threefold molar excess of the dye-NHS was incubated with 1 mg of the antibody for 2 h at room temeprature with continuous rotation. Free dye was removed using a PD SpinTrap G-25 Desalting Column (Cytiva). Protein concentration and degree of labelling (fluoro-to-protein ratio) were determined using NanoDrop spectrophotometric absorbance values. All antibody conjugations yielded a degree of labelling of 2–3 dyes per antibody.
Immunofluorescence of infected cells and RSV virions on glass
Infected cells and uninfected control cells were fixed with 4% paraformaldehyde (043368.9M, Thermo Fisher Scientific) for 10 min at room temperature at experimental end point. All fluorescence of genetically encoded fluorescent proteins was maintained following this fixation and antibody staining protocol. If virions attached to the extracellular membrane of cells needed to be stained, RSV glycoprotein F antibody (11-2-F3; referred to as anti-RSV F; DyLight550, NBP2-50412R and Alexa Fluor 488, NBP2-50412AF488, NOVUS Biologicals) staining was carried out before permeabilization and blocking. Antibody diluted to the appropriate concentration in PBS + 10% FBS was added to cells and incubated for 1 h at room temperature (1:500 anti-RSV F (AF488/DL550)). Following incubation, samples were washed three times in PBS. Samples were subsequently blocked and permeabilized, simultaneously, with blocking buffer (PBS + 10% FBS + 0.05% Triton X-100) for 1 h at room temperautre. Antibodies and recombinant proteins were diluted to the appropriate concentration in blocking buffer and were incubated for 1 h at room temperature or overnight at 4 °C (for recombinant proteins: 1:1,000 of 0.4 mg ml−1 DARPin-P-mRuby3, 1:1,000 of 0.4 mg ml−1 DARPin-P-sfGFP, 1:500 of 0.4 mg ml−1 DARPin-P; for antibodies: 1:1,000 (for ExM, 1:100) of 0.5 mg ml−1 anti-RSV N (AF555/AF647), 1:1,000 RSV phosphoprotein antibody (2H102; Janelia Fluor 646; referred to as anti-RSV P (JF646); NBP2-50276JF646, NOVUS Biologicals), 1:10,000 (for ExM, 1:5,000) rabbit anti-P antiserum80 (referred to as anti-RSV P (polyclonal)), 1:1,000 RSV M2-1 protein antibody (5H5; Alexa Fluor 647; referred to as anti-RSV M2-1 (AF647); NBP2-50481AF647, NOVUS Biologicals). Following incubation, samples were washed three times in wash buffer (PBS + 0.05% Triton X-100). If secondary antibody staining was required, the antibodies were diluted to the appropriate concentration in blocking buffer and were incubated for 1 h at room temperature (1:300 of DYKDDDDK tag monoclonal antibody (FG4R; DyLight 488; referred to as anti-FLAG tag-DL488; MA1-91878-D488, Thermo Fisher Scientific), 1:300 of donkey anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody (Alexa Fluor 568; referred to anti-rabbit-AF568; A10042, Thermo Fisher Scientific). Following incubation, samples were washed three times in wash buffer (PBS + 0.05% Triton X-100). Following incubation, samples were washed three times in wash buffer. If whole-cell segmentation was required, total protein was stained using Pacific Blue succinimidyl ester (Thermo Fisher Scientific; 1:2,000 of 1 mg ml−1 stock solution made up in anhydrous DMSO) in PBS for 10 min at room temperature followed by three washes in wash buffer. Samples were kept in PBS at 4 °C until imaging. To visualize extracellular G protein expression on RSV infected cells, 1:1,000 dilution of AbG-fluoro (JF646/JF405) was added directly to the cell culture media and maintained for 30 min at 37 °C. Following incubation, the cells were washed three times in PBS, fixed and stained, as described above.
Virion protein levels were assessed by incubating RSV inoculum in PBS (total volume 100 µl) for 1 h at 37 °C directly in a well of a 96-well square glass-bottom plate. Following incubation, virions were fixed by adding 4% paraformaldehyde directly into the inoculum and incubating for 15 min at room temperature. The antibody staining was carried out as described above. In addition to the antibodies listed above, 1:1,000 FluoTag-X2 anti-ALFA (Atto 488; referred to as anti-ALFA (Atto488); N1502, NanoTag Biotechnologies) was used for virion labelling.
All antibodies, purified proteins and staining protocols used in this study have been optimized to yield minimal to no background foci signal, such that the staining can be used to evaluate RSV infections at a single vRNP resolution. The specific antibody–fluorescent conjugations used per experiment are listed in Supplementary Table 1.
smFISH
A detailed step-by-step protocol for smFISH staining on different samples can be found in Supplementary Information.
smFISH probe generation
Stellaris probe designer (https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer) was used to design probes targeting the entirety and subregions of the negative-sense RSV genome, intergenic regions of the positive-sense RSV antigenome (to minimize cross-reactivity with viral mRNAs), positive-sense RSV transcript mRNAs and SunTag mRNA. For detection of defective viral genomes (DVGs), probes targeting the first 1,000 bp of the RSV 5′ trailer region were used, based on the strong enrichment of copy-back DVG rejoin junctions within this area28,29. This design enables sensitive and robust detection of diverse DVG species, as previously validated by PCR and RNA sequencing methodologies in clinical and experimental samples28,29. Oligonucleotide probes were ordered from Integrated DNA technologies (sequences are listed in Supplementary Table 1). All probes targeting a single RNA were pooled and labelled with ddUTP-coupled ATTO 565 (AD565-31, ATTO-Tec) or ATTO 633 (AD633-31, ATTO-Tec) dyes using terminal deoxynucleotidyl transferase (EP0161, Thermo Fisher Scientific) as previously described81. For the RSV transcriptome, probes targeting all RSV transcripts were pooled. Fluorescent probes were purified by ethanol precipitation and resuspended in nuclease-free water to a final stock concentration of 30 µM.
smFISH of infected cells
The smFISH staining procedure was carried out as previously described82. In brief, following fixation, cells were washed three times in PBS, permeabilized in 100% ethanol for 1 h at 4 °C and then washed three times for 15 min in smFISH wash buffer (2× saline-sodium citrate (SSC), 10% formamide (17899, Thermo Fisher Scientific) in nuclease-free water) at room temperature. Labelled smFISH probes were diluted to 10 nM in hybridization buffer (1% dextran sulfate (D8906, Merck), 2× SSC, 10% formamide, 1 mg ml−1 tRNA (R1753, Merck), 2 mM ribonucleoside vanadyl complex (S1402S, New England Biolabs) and 200 µg ml−1 BSA (AM2616, Thermo Fisher Scientific) in nuclease-free water) and hybridization was performed at 37 °C for 16 h. Unbound smFISH probes were removed by 3 × 30 min washes in smFISH wash buffer at 37 °C and a 15 min wash at room temperature. If whole-cell segmentation was required, total protein was stained using Pacific Blue succinimidyl ester (1:2,000 of 1 mg ml−1 stock solution; P10163, Thermo Fisher Scientific) in PBS for 10 min at room temperature followed by a 15 min wash with smFISH wash buffer at room temperature. Samples were stored at 4 °C and imaged in smFISH imaging buffer (10 mM Tris pH 8, 2× SSC and 0.4% glucose, supplemented with glucose oxidase (G2133, Merck) and catalase (C3515, Merck)). Imaging was performed within 3 days after probe hybridization. The specific smFISH probe sets used in each experiment are listed in Supplementary Table 1.
smFISH of virions and vRNPs on glass
Virion and vRNP vRNA levels were assessed by incubating RSV inoculum in PBS or PBS + 1% Triton X-100 (total volume of 100 µl), respectively, for 4 h at 37 °C directly in a well of a 96-well square glass-bottom plate. Following incubation, samples were fixed by adding 4% paraformaldehyde directly into the inoculum and incubating for 15 min at room temperature. smFISH staining was carried out as mention above with minor modifications aimed at enhancing accessibility for the smFISH probes to bind to the vRNA. Following fixation and washing, vRNPs were permeabilized in 80% ethanol overnight at 4 °C and then washed once for 15 min in smFISH wash buffer at room temperature. Samples were incubated in 80% formamide for 40 min at 37 °C followed by 2 × 15 min washes in smFISH buffer at room temperature. Samples were incubated with 1:2,000 proteinase K in 2× SSC (124568, Merck) for 1 h at room temperature and washed 2 × 15 min in smFISH buffer at room temperature. Probe hybridization and subsequent steps in the protocol were carried out as above.
Combining smFISH and N antibody staining
To combine smFISH with anti-RSV N antibody staining, the wash buffer was additionally supplemented with 3% BSA and the fluorescently conjugated antibody was directly added into the smFISH hybridization buffer at a 1:100 dilution. The remaining protocol is carried out as detailed above.
Combined smFISH and STAb staining
To combine smFISH with STAb–sfGFP staining, after the three washes with smFISH wash buffer at 37 °C, samples were incubated with purified scFv–sfGFP–STrepII16 (STAb–sfGFP; 1:100 dilution in PBS + 10% BSA) for 1 h at room temperature. Following incubation, samples were washed three times in PBS. A final room temperature wash step of 30 min in smFISH wash buffer was then performed before sample storage and imaging in smFISH imaging buffer.
Combining smFISH staining with fluorescence originating from transgenic cell lines
For the Pexo-fluoro cell line, used to visualize all RSV vRNPs, the foci signal (in the AausFP1 fluorescent protein) was well retained following the smFISH protocol and can be visualized simultaneously with the smFISH foci. For the DARPin-P visualizing transgenic cell line, the foci signal (in the BFP fluorescent protein) was somewhat compromised following the smFISH protocol and as such needed to be first imaged post-fixation, but before ethanol permeabilization, and then followed by the smFISH protocol. When DARPin-P signal visualization with smFISH signals were required, experiments were only carried out in the Pexo-fluoro and DARPin-P-fluoro combined transgenic line. Acquiring the Pexo signal both in the post-fixation and post-smFISH imaging rounds allowed it to be used to align the data from the two acquisition rounds, ensuring that accurate values for the DARPin-P signal (from post-fixation imaging) could be used for the cells being analysed.
ExM
ExM was performed as previously described32 with some adaptations. Stained samples were crosslinked with 1.2% paraformaldehyde (11586711, Thermo Fisher Scientific)–3% acrylamide (A4058, Sigma-Aldrich) in PBS for 3 h, washed three times with PBS and incubated for 5 min in monomer solution (1× PBS, 2 M NaCl, 2.5% (w/w) acrylamide, 0.15% (w/w) N,N′-methylenebisacrylamide (M1533, Merck) and 8.625% (w/w) sodium acrylate (408220, Sigma-Aldrich)). Coverslips were placed on top of a drop of 90 μl of freshly prepared gelation solution (monomer solution supplemented with 0.2% (w/w) TEMED (T9281, Sigma-Aldrich) and 0.2% (w/w) APS (A7460, Sigma-Aldrich)) and incubated for 1 h at 37 °C. Gels were then incubated in digestion solution (8 units per millilitre proteinase K (1.24568, Merck), 1× TAE, 0.5% Triton X-100 (X100, Sigma-Aldrich) and 0.8 M guanidine HCl (G3272, Sigma-Aldrich) for 3 h at 37 °C and washed in PBS containing DAPI (1 µg ml−1). Expansion was performed by several washings with excess volume of Milli-Q water (approximately 4.5-fold expansion). Expanded samples were immobilized on 25 mm (no. 1.5H) coverslips covered with 0.01% (w/v) poly-L-lysin (P8920, Sigma-Aldrich) and imaged.
Flow cytometry-based assessment of successful RSV infections
A flow cytometry-based assay quantifying expression of viral G protein by infected cells was used as a readout of successful RSV infections. This assay was used to assess RSV infection success in the DARPin-P-fluoro transgenic A549 cell line compared with WT A549 cells. WT A549 cells and DARPin-P-fluoro cells were plated, in parallel, on a 96-well glass-bottom imaging plate (for establishment of MOI) and a 24-well plate (for flow cytometry assessment). The next day, cells were infected with RSV (0.001 PFU per cell). For MOI establishment, following 6 h of infection, the cells plated on the imaging plate were fixed and subject to antibody staining for anti-RSV N and F and the number of infected cells (N+/F−) determined to obtain a MOI. For flow cytometry assessment, at 6 h, cells infected for flow cytometry assessment were treated with an RSV fusion inhibitor (TMC353121) to prevent subsequent infections. Flow cytometry assessment was carried out at 24 h and 48 h following viral inoculum addition. An hour before flow cytometry, infected cells (and uninfected control cells) were incubated with AbG-fluoro (1:1,000, in cell culture media) for 45 min at 37 °C. After antibody incubation, cells were rinsed in PBS, trypsinized, fixed (in suspension, with 4% paraformaldehyde for 10 min at room temperature with constant rotation) and suspended in PBS. Flow cytometry analysis was performed using Cytoflex S (Beckman Coulter) and CytExpert software (see Supplementary Fig. 1). The percentage of successful infections in each cell line was calculated in relation to the fraction of infected cells at 6 h post-inoculum addition (MOI) to determine whether expression of the DARPin-P-fluoro reduced the efficiency of RSV infection success.
Microscopy
Microscope and image acquisition details
All fluorescence microscopy was carried out on a Nikon Ti2 inverted microscope equipped with a Yokagawa CSU-X1 spinning disc and a Prime 95B sCMOS camera (Photometrics), unless stated otherwise. Imaging was performed using a ×60/1.40 NA oil-immersion objective. Image acquisition was performed using NIS Elements software, making use of the ‘perfect focus system’ to correct for Z drift during timelapse imaging experiments. The microscope was equipped with a temperature-controlled incubator, and imaging for live-cell experiments was performed at 37 °C and for fixed-cell experiments at room temperature. For timelapse imaging experiments, x, y positions for imaging were randomly selected. Position selection was carried out immediately following viral inoculum addition and before observing foci in infected cells. Images were acquired every 5, 10, 20 or 30 min (exact intervals are recorded in the figure legends) for 6 h (short-term timelapse imaging) or 12–48 h (for overnight and longer-term timelapse imaging), using 50–70-ms exposure times (50 ms for the BFP channel and 70 ms for all other channels). Multiple Z-slices (approximately 9–15 slices with a 0.8-µm step size) were imaged for each channel, ensuring that the entirety of the cell was captured. For fixed cell analysis of immunofluorescence or smFISH staining, x, y positions for imaging were selected either randomly using the cell segmentation channel (when data regarding the frequency of a phenotype were assessed) or targeted using the channel of interest (when the frequency of a phenotype was not assessed). For smFISH staining in which the transgenic cell line DARPin-P-fluoro signal was acquired post-fixation, the same x, y positions are used to acquire the smFISH staining. Images were acquired at 45% laser power and 70-ms exposure (for all lasers) with a 0.8-µm step size and a Z-stack slice number that ensures the entirety of the cell is captured.
For high temporal resolution imaging of vRNP dynamics following RSV infection, real-time imaging was performed on a Nikon Ti2 inverted microscope equipped with a Yokogawa CSU-X1 spinning disk confocal unit and a Kinetix sCMOS LFOV camera (Photometrics). A Nikon Plan Apo λD ×60/1.42 NA oil-immersion objective was used. Excitation was provided by a 488-nm laser line (50% AOTF power) coupled with a 405/488/555/640/730 RPC-UF1 dichroic and a 525/50 emission filter. Timelapse acquisition was conducted for 4 h at a 10-s interval with the Ti2 perfect focus system engaged for Z-drift correction. Camera settings included 16-bit mode, no binning and 250-ms exposure time. The large 29.4-mm diagonal sensor of the Kinetix camera allowed an expanded field of view (approximately 320 µm × 320 µm), increasing imaging throughput. Fast z-stack acquisition was enabled using the NIDAQ Piezo. Image acquisition and hardware control were managed with NIS Elements software.
ExM datasets were acquired on a Zeiss LSM900 microscope equipped with an Airyscan2 detector using a ×63/1.3 NA objective. Images were collected in Multiplex SR-2Y mode as z-stacks with 0.120-µm spacing. Excitation was performed using 640-nm (20% laser power), 561-nm (2% laser power), 488-nm (30% laser power) and 405-nm (20% laser power) laser lines.
Post-acquisition data processing and video generation
Maximal intensity projections for all Z-slices (where multiple Z-slices were acquired) were generated using NIS Elements software (Nikon) and all downstream analyses were performed on these projections.
ExM image stacks were deconvolved in Huygens Professional (Scientific Volume Imaging, v20.10) using the classic maximum likelihood estimation algorithm with a theoretical point spread function (PSF), allowing up to 40 iterations.
The single-object-grid tool (https://github.com/TanenbaumLab/SingleObjectGrid) was used to crop a small field of view around tracked cells to create a stabilized time series with the cell centroid centred. The napari-animation plugin was used to generate video output from the napari viewer to create supplementary videos (https://napari.org/napari-animation/).
Quantification
Foci quantification
For all analyses, infected cells were manually identified by the presence of RSV vRNPs (as determined by the Pexo foci, anti-RSV N antibody staining or (−)vRNA smFISH staining). Only cells that were completely in the field of view were included in the analysis.
For fixed-cell analysis, infected cells were manually segmented in ImageJ, and foci detection and foci integrated intensity quantification were performed using the ‘detect particles and colocalization analysis’ function in the ComDet (v0.5.4) plugin in ImageJ (https://github.com/UU-cellbiology/ComDet). Plugin spot detection parameters were optimized for each type of foci analysed. The intensity thresholds used in the analyses were established for each individual experiment using the uninfected control samples. The specific parameters used were: smFISH viral N mRNA spot detection; approximate particle size of 3 µm, intensity threshold range of 15–30 or more (that is, 15–30 times or more the signal-to-noise ratio), large particles not included; smFISH viral genome and antigenome spot detection; approximate particle size of 4 µm, intensity threshold range of 5–50 or more, large particles included; transgenic cell line-derived and antibody staining-derived RSV vRNP spot detection; approximate particle size of 4 µm, and intensity threshold range of 5–200 or more, large particles included. For individual vRNP spot detection (including smFISH, transgenic cell line and antibody staining-derived foci), an additional filtering step was introduced, in which only spots that had an N area value > 6 and value < 30 (pixels in foci) were included in the analysis. Intensity-based cut-offs for each channel were used to call colocalization. These cut-off values were established for each individual experiment using both the uninfected control sample and the individual channel of interest. The lowest intensity where a foci was detected (individual channel of interest) above background was used as the intensity cut-off to call positive and negative values. Foci integrated intensity values (sum of all pixels intensities inside the segmented spot area minus the average spot-specific backgrounds). Background value, calculated as the average intensity of pixels along the perimeter of a bounding rectangle surrounding the segmented spot, of less than 0 was converted to 0 to aid with analysis and data visualization. Integrated intensity values were normalized (to the median value of DARPin-P− foci, or to the maximum intensity value) to allow for optimal data comparison. For antibody staining experiments, only genomes that were N+/F− were considered to be inside cells. If the Pexo-fluoro transgenic cell line was utilized, only Pexo+ foci were considered to be vRNPs inside infected cells.
Quantification of the number of foci per virion was carried out manually. Pexo-fluoro cells were infected with an inoculum containing RSV (low MOI, exact value recorded in figure legends) and either the viral polymerase inhibitor PC786 or the translation inhibitor emetine to prevent infection progression. If timelapse data were acquired, imaging was started immediately following inoculum addition. The start of infection was defined as the first time point a Pexo-fluoro foci was observed. Only cells in which a minimum of two time points before viral entry were observed were included in the analysis. Foci number was determined for each time point until the end of the acquisition. Only cells that remained completely within the field of view for the entire duration of analysis were included. In addition, only infected cells in which minimally 10 h of infection was observed were used for downstream analysis. In cases in which foci number per virion was assessed at a fixed time point, 1 h after inoculum addition, the fusion inhibitor TMC353121 was added to prevent subsequent infections. Three hours after fusion inhibitor addition (4 h post-inoculum addition), the cells were imaged. The number of foci in each infected cell was quantified.
Quantification of the time foci appeared and the number of foci present at each time frame in live-cell timelapse imaging videos was carried out manually. Only cells that remained completely within the field of view for the entire duration of the analysis were included. For experiments in which infection was followed from the moment of viral cell entry, only cells in which a minimum of two time frames before viral entry were observed were included. For vRNP analysis, only foci that were present for a minimal of two consecutive time frames were considered in the analysis to minimize false foci calling. For assessing the DARPin-P state, infections were classified as DARPin-P+ if one or more of the infecting vRNPs were DARPin-P+ even if the cell additionally had DARPin-P− vRNPs. Quantification of the number of translating viral reporter mRNAs per cell (infection with the upstream and downstream SunTageng RSV strains) was performed, based on previously described guidelines16. In brief, the number of translation sites at each individual time frame were assessed until individual translation sites could no longer be detected (high translation rate leading to the depletion of free cellular STAb due to the cytoplasmic accumulation of mature SunTag peptides)53. AbG-fluoro signal accumulation was also assessed manually and recorded at the first time frame positivity could be observed. Foci calling (Pexo and DARPin-P signal for vRNPs and translation sites for the SunTageng RSV downstream engineered strain) and AbG-fluoro accumulation calling were independently validated by another laboratory member. If intensity quantification of foci in timelapse videos was performed, the cell of interest was manually segmented at each time point and foci intensity quantified using the ComDet plugin. To facilitate visual inspection of general trends in heterogenous intensity values over time (for individually tracked foci), moving averages were plotted with a sliding window of three time points.
Quantification of viral transcription rate
As the number of translating SunTag mRNAs closely correlated with the number SunTag mRNAs (Extended Data Fig. 7f), the increase in the number of translating SunTag mRNAs over time was used as a proxy to calculate viral transcription rate. The number of viral mRNA translation sites within a 1-h period as initial observation was plotted against time. Graphs for individual infections were fit with a linear function using GraphPad prism (v10.1.0) and the slope of this function was used as the viral transcription rate.
Quantification of ExM datasets
Quantification was performed using Imaris software (Bitplane v9.7.2). vRNPs were segmented based on the polyclonal-RSV P antibody signal using the Surface function with a grain size of 100 nm and background subtraction (maximum sphere diameter of 300 nm). All images within the same experiment were processed using identical threshold values, and background surfaces were defined from the inverse portion of the intensity histogram. The mean DARPin-P intensity per segmented vRNP was corrected by subtracting the corresponding background mean and subsequently normalized to the average intensity across all particles from the same experiment. A cut-off value of 0.3 was applied to classify viral particles as DARPin-P+ or DARPin-P− (Extended Data Fig. 8a). Particle size was defined as the length of the longest axis of the bounding box aligned to the object. Statistical analyses were performed using Prism 10 (GraphPad Software).
Proteomic quantitative analysis
Protein and phospho-site identification and quantification were performed using Andromeda search engine implemented in MaxQuant (v2.3.0)83. Spectra were searched against human proteomes downloaded from Uniprot (on 22 August 2022), RSV custom proteome derived from human RSV subgroup A, strain Long (GenBank accession: AY911262) and DARPin-P sequence. False discovery rate was set at 1% for both peptide and protein identification. MaxQuant search was performed with ‘match between run’ activated in the searches. Phosphorylation on STY residues was set as a variable modification in addition to protein N-terminal acetylation and methionine oxidation. All other parameters were set to default values. MaxQuant (proteinGroups) outputs were used for downstream relative quantification analysis. Proteins highlighted as potential contaminant and reverse were filtered out. R-package limma (v3.52.4)84 was used for statistical analysis for protein intensities, applying a moderated t-test, with P values adjusted for multiple testing using Benjamini–Hochberg methodology.
Statistical analysis
All statistical analyses performed using GraphPad prism (v10.1.0, GraphPad Software) and Microsoft Excel. Unless stated otherwise, statistical tests were performed using a P value of 0.05 as a cut-off for significance and assuming normal distribution of experimentally determined averages. The type of test and the type of error bars used in figures are indicated in the figure legend or in the figure itself. The n values are recorded in the figures and included in Supplementary Table 1. In addition, Supplementary Table 1 presents an overview of the number of experimental repeats, the total number and type of observations per condition, as well as situations in which the same raw data have been used for multiple independent analyses. Graph creation was also performed in GraphPad.
Material availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact M.E.T. ([email protected]). The unique or stable reagents generated in this study are available from the lead contact with a completed Material Transfer Agreement. The plasmids generated in this study for the vRNP visualization tools (Pexo–SunTag, Pexo–ALFATag, M2-1exo–SunTag, Nexo–SunTag and DARPin-P–BFP) have been deposited in Addgene.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.