Mice
Previously published genetically engineered mouse strains were used in this study: KrasLSL-G12D/+ (ref. 19), Trp53flox/flox (ref. 62), KrasFSF-G12D/+ (ref. 21), Trp53frt/frt (ref. 26), Rosa26mTmG/+ (ref. 36); and Hipp11FSF-GGCB, Hipp11FSF-BG, Slc4a11FSF-MCD and HopxFSF-MACD reporters were generated in this study as described in detail below. All mice bearing autochthonous KP lung tumours were maintained in a C57BL/6 × Sv129 mixed background. NOD.Cg-Prkdcscid;Il2rgtm1Wjl/SzJ(NSG)63 mice (The Jackson Laboratory, 005557) were used as recipients in all allotransplant studies. All mice were monitored by the investigators and veterinary staff at the Research Animal Resource Center at Memorial Sloan Kettering Cancer Center (MSKCC) and housed under a 12 h–12 h light–dark cycle at 20–25 °C and 30–70% humidity with food and water provided ad libitum.
Autochthonous and transplantation models of lung cancer
Autochthonous LUAD tumours were induced in KrasLSL-G12D/+;Trp53flox/flox or KrasFSF-G12D/+;Trp53frt/frt (KPfrt) mice with 1 × 108–1 × 109 plaque-forming units (PFU) of AdSPC-Cre, AdSPC-FlpO (Iowa Viral Vector Core), or lentiviral FlpO at 3 × 105 or 6 × 105 transforming units, as previously described64, in mice that were aged between 8 and 12 weeks. Immunocompromised NSG mice were used as recipients for either subcutaneous, orthotopic or intravenous transplantation of KP LUAD cell line allografts. For subcutaneous transplantation, cells were resuspended in S-MEM (Gibco, 11380-037) and mixed with Matrigel (Thermo Fisher Scientific, CB-40230C) at a 1:1 ratio. Then, 250,000 cells were implanted subcutaneously into both flanks of NSG mice. For orthotopic transplantation, sorted cells were resuspended in PBS (Gibco, 10010-023) and intratracheally administrated to NSG mice. For intravenous transplantation, 200,000 cells were resuspended in S-MEM and injected into NSG mice through the tail vein. All cell lines were continuously monitored for mycoplasma contamination. Approximately equal numbers of male and female mice were included in all experimental groups in all mouse experiments. Mice were treated in accordance with all relevant institutional and national guidelines and regulations, and mice were euthanized by CO2 asphyxiation, followed by intracardiac perfusion with S-MEM to clear tissues of blood when appropriate. A complete list of mice along with age, sex and age of tumour used in experiments is available (Supplementary Table 4). All animal studies were approved by the MSKCC Institutional Animal Care and Use Committee (protocol 17-11-008). Sample sizes were determined based on our previous experience with similar models rather than statistical methods. We found this sufficient to detect biologically meaningful differences while minimizing animal use; experiments were randomized when feasible. Blinding was not possible as treatment effects on tumour volume were readily distinguishable between groups. Tumour burden limit was defined as a single tumour >2 cm in diameter, tumour volume >10% of body mass or multiple tumours with a cumulative volume >3,000 mm3. These limits were not exceeded in any of our experiments.
Generation of donor vectors for embryonic stem cell targeting
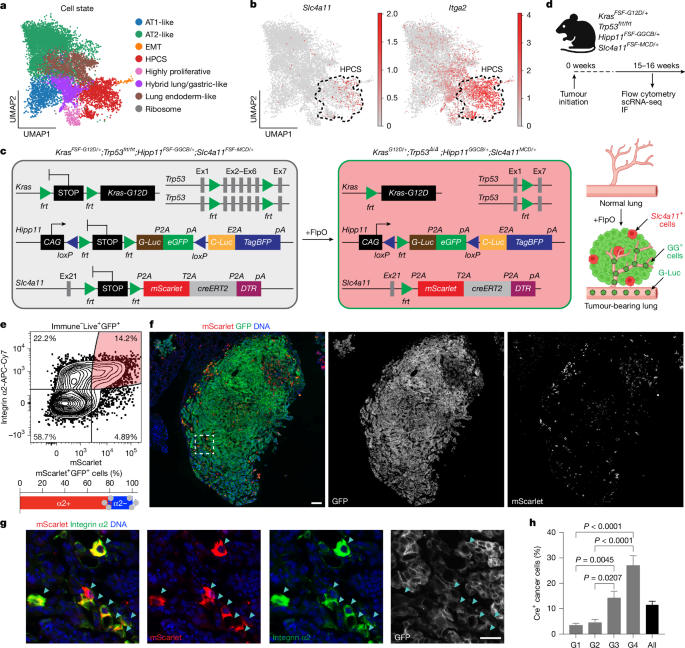
For the generation of the Slc4a11-FSF-MCD donor vector, homology arms of around 1,200 bp in length 5′ and 3′ to the end of Slc4a11 exon 21 (Extended Data Fig. 1b) were amplified from genomic DNA of C57BL/6 mES cells using high-fidelity PCR (NEB, M0494). A homology-directed repair template donor vector was constructed by flanking the frt-bGlobinpA-(PGK-Hygromycin-pA)i-frt-P2A-mScarlet-T2A-CreERT2-P2A-DTR-WPRE-bGHpA cassette with the 5′ and 3′ homology arms and cloned into the pUC19 plasmid backbone (Takara Bio, 638949) using Gibson assembly (NEB, E2611).
For the generation of the Hipp11-FSF-GGCB donor vector, homology arms of around 5,000 bp in length 5′ and 3′ to the safe harbour of Hipp11 intergenic region (positioned between the Eif4enif1 and Drg1 genes; Extended Data Fig. 1e) were amplified from genomic DNA of C57BL/6 mES cells using high-fidelity PCR. A homology-directed repair template donor vector was constructed by flanking the CAG-loxP-frt-Neomycin-PGKpA-SV40pA-frt-G-Luc-P2A-meGFP-bGlobinpA-loxP-C-Luc-E2A-TagBFP-3xFlag-WPRE-bGHpA (GGCB) cassette with the 5′ and 3′ homology arms and cloned into the pUC19 plasmid backbone using Gibson assembly.
For the generation of Hopx-FSF-MACD donor vector, homology arms of around 1,500 bp in length 5′ and 3′ to the end of Hopx exon 3 (Extended Data Fig. 6a) were amplified from genomic DNA of C57BL/6 mES cells using high-fidelity PCR. A homology-directed repair template donor vector was constructed by flanking the frt-bGlobinpA-(PGK-Hygromycin-pA)i-frt-P2A-mScarlet-AkaLuc-T2A-CreERT2-P2A-DTR-WPRE-bGHpA cassette with the 5′ and 3′ homology arms and cloned into the pUC19 plasmid backbone using Gibson assembly.
Validation of the Hipp11
GGCBreporter
To validate the functionality of the GGCB cassette, we performed ex vivo transformation of AT2 cells isolated from a KPfrt;Hipp11FSF-GGCB/+ chimeric mouse using lentiviral vectors encoding either codon-optimized Flp recombinase (flpO) alone or flpO linked to creERT2. In these experiments, FlpO activates oncogenic KRAS(G12D), deletes Trp53, and initiates expression of the GG cassette, whereas subsequent activation of CreERT2 with 4-hydroxytamoxifen (4-OHT) results in a switch from GG to CB (Extended Data Fig. 1h–k). G-Luc activity was increased 13 days after transformation in all conditions (Extended Data Fig. 1i (top)), whereas C-Luc activity was observed only after 4-OHT stimulation in organoids transduced with the vector encoding both flpO and creERT2 (Extended Data Fig. 1i (bottom)). Moreover, we performed flow cytometry and fluorescence imaging analyses on organoids under these four conditions. We found that eGFP was expressed at the baseline following flpO and a switch to TagBFP was observed only after 4-OHT exposure in the organoids transduced with both flpO and creERT2 (Extended Data Fig. 1j,k). Similar results were obtained in subcutaneous transplants (Extended Data Fig. 2a–c) and autochthonous lung tumours (Extended Data Fig. 2d–g) in vivo, both by detection of G-Luc and C-Luc from repeated blood samples and by fluorescence imaging of tumours at the end point.
Embryonic stem cell targeting, genotyping and chimera generation
A KrasFSF-G12D/+;Trp53frt/frt (KPfrt) mES cell line in the C57BL/6J background was generated by crossing a hormone-primed C57BL/6J Trp53frt/frt female mouse with a KrasFSF-G12D/+;Trp53frt/frt male mouse. At 3.5 days after coitum, blastocysts were flushed out from the pregnant uterus, isolated and cultured on a mouse embryonic fibroblast (MEF) feeder layer. Individual ES cell lines were genotyped by PCR detection of KrasFSF-G12D/+, Trp53frt/frt and Zfy (Y-chromosome-specific).
For the generation of Slc4a11FSF-MCD/+, HopxFSF-MACD/+ and Hipp11FSF-GGCB/+ knock-in mES cells, donor vectors (Slc4a11-FSF-MCD, Hopx-FSF-MACD or Hipp11-FSF-GGCB, respectively) and ribonucleoprotein (RNP) complex containing HiFi Cas9 nuclease (IDT, 1081061) and crRNA–tracrRNA duplex (IDT) were co-transfected into the KPfrt mES cell line by electroporation (Lonza, 4D Nucleofector). A list of the sequences of crRNAs is provided in Supplementary Table 4.
KPfrt mES cells were thawed 2 days before targeting and the media were changed 1 day and 2 h before electroporation. Before electroporation, sequence-specific crRNA and universal tracrRNA were resuspended in IDTE buffer (IDT) at a concentration of 200 µM and the crRNA–tracrRNA duplex was then formed (final concentration, 44 µM) by combining an equimolar concentration of crRNA and tracrRNA and annealing at 95 °C for 5 min (followed by cooling down to room temperature at ramp rate of 0.1 °C s−1). RNP complexes were formed by combining 22 pmol of crRNA–tracrRNA duplex and 22 pmol HiFi Cas9 nuclease and incubating at room temperature for 20 min. For each electroporation, 500,000 mES cells depleted of MEFs, 1 µl donor vector (3 µg µl−1), 1 µl RNP complex, 2 µl electroporation enhancer (10 µM, IDT), 16.4 µl Nucleofector P3 primary cell solution and 3.6 µl Nucleofector Supplement 1 were combined and loaded into electroporation cuvette. The ES cells were then plated onto the top of feeder MEFs and, 48 h later, the ES cells were selected with either hygromycin (Slc4a11-FSF-MCD and Hopx-FSF-MACD, 150 µg ml−1) or G418 (Hipp11-FSF-GGCB, 400 µg ml−1) for 1 week. Resistant clones were manually picked, expanded and validated by genotyping using the primers listed in Supplementary Table 4.
Generation of genetically engineered reporter mouse strains
Chimeric F0 mice were obtained by injecting genotype-verified mES cells into host embryos at the eight-cell stage and genotyped at 2 weeks of age. F0 mice were crossed into the KrasFSF-G12D/+;Trp53frt/frt background to generate mice appropriate for the given experiments.
Generation of LUAD reporter and lineage-tracing cell lines
For the generation of the Slc4a11MACD/+;KP LUAD reporter cell line, a KP LUAD cell line derived from a mouse bearing autochthonous KrasLSL-G12D/+;Trp53flox/flox tumours at 24 weeks PTI was generated. The KP LUAD cells were then co-transfected with the Slc4a11-FSF-MACD donor vector together with the U6-sgSlc4a11-EFS-Cas9 vector expressing guide RNA (ACATATGGGGAGGTATGAGC) targeting the last exon of Slc4a11 at a 1:1 ratio using Lipofectamine 3000 (Thermo Fisher Scientific, L3000015). The transfected cells were selected with hygromycin (Sigma-Aldrich, 400053) at a concentration of 150 µg ml−1 for 2 weeks and single-cell-derived drug-resistant clones were manually picked for expansion and genotyping with the following primers (5′ KI_F1 and 5′ KI_R1; 3′ KI_F1 and 3′ KI_R1). The single-cell-derived clones were transduced with AdCMVFlpO (Iowa Viral Vector Core) at a multiplicity of infection (MOI) of 500 to remove the frt-bGlobinpA-(PGK-Hygromycin-pA)i-frt STOP cassette. Excision of the STOP cassette was confirmed by genotyping spanning the left homology arm and mScarlet (recombined; 5′ KI_F1 and 5′ KI_R2) and by flow cytometry analysis detecting mScarlet fluorescence (Extended Data Fig. 9c,d). An additional Slc4a11-MACD;KP LUAD reporter cell line was generated through ex vivo transformation of an AT2 organoid culture intermediate, as described previously9 (see below). The lentiviral lineage tracing vectors (Lenti-EFS-Flex-TagBFP-PGK-eGFP or PGK-Gluc-miRFP670-EFS-lox-BFP-lox)9 (Extended Data Fig. 9g–i) were transduced into the Slc4a11-MACD;KP LUAD reporter cell lines and sorted using fluorescence-activated cell sorting (FACS) based on eGFP or miRFP670 fluorescence. KPfrt;HopxMACD/+ LUAD reporter cell lines were generated as previously described9 or derived from autochthonous KrasFSF-G12D/+;Trp53frt/frt;HopxMACD/+;Hipp11BG/+ tumours at 20 weeks PTI, in which the Hipp11BG/+ allele enables switching from baseline (TagBFP+) to lineage-traced (GFP+) fluorescence. All cell lines tested negative for mycoplasma contamination.
Dissociation of LUADs and lung tissue
For isolation of normal AT2 cells and autochthonous LUAD cells, mice were euthanized at the indicated timepoints after tumour induction and were perfused with sterile S-MEM (Gibco, 11380-037) through the right ventricle of the heart. Dissected lungs or microdissected tumours were dissociated with a mixture of dispase II (Corning, 354235, 0.6 U ml−1), collagenase type IV (Thermo Fisher Scientific, 17104019, 167 U ml−1) and DNase I (StemCell Technologies, 07469, 10 U ml−1) in S-MEM solution at 37 °C as previously described4 for 1 h. The dissociated cells were filtered using a 100 µm filter and centrifuged at 1,500 rpm for 10 min at 4 °C. The supernatant was removed by aspiration and red blood cell lysis was performed using BD Pharm Lyse (BD Biosciences, 555899) for 1 min on ice. Cells were then washed with sterile medium containing 2% heat-inactivated FBS (Hyclone, SH30910.03), passed through a 40 µm filter and pelleted at 300g for 5 min at 4 °C. The supernatant was removed, and live cells were purified using the Akadeum Dead Cell Removal Microbubble kit according to the manufacturer’s instructions (Akadeum Life Sciences, 11510-211). Cells were resuspended in FACS buffer (2% heat-inactivated FBS in PBS) and counted for use in FACS, as described below.
Flow cytometry analysis and FACS
Cells were prepared as described above, and Fc block (BD Biosciences, 553142) was added on ice for 10 min before staining with the appropriate antibody panel (Supplementary Table 4). After 20 min of staining on ice, cells were washed twice with FACS buffer and pelleted by a 5 min spin (300g at 4 °C). The cell pellets were resuspended in PBS with 2% heat-inactivated FBS containing DAPI (Sigma-Aldrich, D9542, 1 μg ml−1) or Helix NP NIR (BioLegend, 425301, 5 nM) to identify dead cells. Cell sorting was performed at the Flow Cytometry Core Facility at Sloan Kettering Institute/MSKCC, using a BD FACS Aria Sorter. Cells were sorted using the 4-way purity mode. Cancer cells were sorted as (CD45/CD31/CD11b/CD11c/F4/80/TER-119)−/Helix NP NIR− or DAPI− (live); the specific fluorescence-positive cell populations are indicated in each experiment.
Alveolar organoid culture and ex vivo transformation protocol
FACS-purified AT2 cells (gated as MHCII+EPCAM+SCA1−podoplanin−lineage−(CD45/CD31/CD11b/CD11c/F4/80/Ter-119)−DAPI−) from KPfrt;Hipp11GGCB/+ chimeras were transduced by lentivirus (Lenti-PGK-FlpO or Lenti-PGK-FlpO-P2A-creERT2) at a MOI of 10 by spinfection (600g, 37 °C, 30 min). Then, 4,000 transduced AT2 cells were resuspended with 50,000 primary pulmonary endothelial cells isolated from 4-week-old Rosa26mTmG/+ mice by FACS (CD31+CD45−DAPI−) in 50 µl alveolar organoid culture medium (Ham’s F-12 (Thermo Fisher Scientific, 11765047), 10% FBS (Hyclone, SH30910.03), 1% GlutaMAX (Thermo Fisher Scientific, 35050061), 1% penicillin–streptomycin (Thermo Fisher Scientific, 15070063), 1% ITS (Millipore Sigma, I3126) and 1% HEPES (Thermo Fisher Scientific, 15630080)). The resuspension was then mixed with 50 µl Matrigel (Thermo Fisher Scientific, CB-40230C) and placed into cell culture inserts (Thermo Fisher Scientific, 08-770). Alveolar organoid culture medium (500 µl) was added to the reservoir (Thermo Fisher Scientific, 353504) outside the insert and replaced every 3 days. Primary organoids were digested at day 7 for secondary organoid culture with 5 U ml−1 dispase (Corning, 354235) for 1 h at 37 °C and replating without endothelial cells to select for transformed tumour spheres. 4-OHT (Sigma-Aldrich, H6278) was added at a concentration of 1 µM at day 10. The supernatants were collected every 3 days for G-Luc and C-Luc measurement starting at day 1. Organoids were imaged using the EVOS M5000 microscope and dissociated for flow cytometry analysis at day 16 (6 days after 4-OHT).
IF and immunohistochemistry
Mice were euthanized by CO2 asphyxiation followed by systemic perfusion with S-MEM (Gibco, 11380-037) or PBS (Gibco, 10010-023) to clear lungs of blood. Tissues were fixed in 10% neutral-buffered formalin (Sigma-Aldrich, HT501128) for 24–48 h at 4 °C and either embedded in paraffin or dehydrated using 30% sucrose for 16–24 h before embedding in OCT compound (Thermo Fisher Scientific, 23-730-571) at −80 °C.
IF imaging was performed on 5 µm formalin-fixed paraffin-embedded (FFPE) sections or 7 µm cryosections. FFPE sections were deparaffinized and heat-induced antigen retrieval was performed using EDTA antigen retrieval buffer (Sigma-Aldrich, E1161). For cryosections, the slides were air-dried for 1 h at room temperature and fixed in acetone at −20 °C for 10 min. The sections were blocked in donkey immunomix (0.2% BSA (Sigma-Aldrich, 810533), 5% donkey serum (Thermo Fisher Scientific, 31874), 0.3% Triton X-100 (Thermo Fisher Scientific, BP151-100) in PBS (Gibco, 10010-023)) at room temperature for 30 min. Incubation of primary antibodies against GFP (Abcam, ab13970), integrin α2 (Abcam, ab181548), Ki-67 (Thermo Fisher Scientific, 14-5698-82), pan-cytokeratin (Agilent Dako, M351529-2), uPAR (R&D Systems, AF807), Flag (Sigma-Aldrich, F1804), NKX2.1 (Abcam, ab76013), HMGA2 (Cell Signaling, 8179), SPC (Sigma-Aldrich, AB3786), HOPX (Santa Cruz Biotechnology, sc-398703), Cre recombinase (Cell Signaling, 15036) and HNF4α (Cell Signaling, 3113) diluted in donkey immunomix was performed at 4 °C overnight. AlexaFluor-conjugated secondary antibodies raised in donkey were used for signal detection (Invitrogen, A31573, A78948, A10037, A78947). Sections were counterstained with 1 µg ml−1 DAPI (Sigma-Aldrich, D9542) for 10 min and mounted with coverslips using Mowiol mounting reagent (EMD Millipore, 475904). Mounted slides were imaged using the Zeiss Axio Imager Z2 and ZEN 2.3 software or digitally scanned using Mirax Midi-Scanner (Carl Zeiss AG). Image analysis was performed using Fiji software.
Haematoxylin and eosin staining was performed using a standard protocol and tumour grades were assigned using an AI-based Aiforia software (NSCLC_v25 algorithm, Aiforia Technologies), as described previously65. For immunohistochemistry, tissue sections were incubated at 58 °C for 1 h and loaded onto the Leica Bond RX and dewaxed at 72 °C, followed by antigen retrieval using EDTA-based ER2 solution (Leica, AR9640) at 100 °C for 20 min. Primary antibodies against Cre recombinase (Cell Signaling Technology, 15036), uPAR (R&D Systems, AF543), NKX2-1 (Abcam, ab76013) or HMGA2 (Cell Signaling Technology, 8179) were incubated at room temperature for 1 h, followed by an 8-min incubation with the Leica Bond Polymer anti-rabbit or anti-goat HRP reagent (Polymer Refine Detection Kit, Leica, DS9800). The mixed DAB reagent (Polymer Refine Detection Kit) was applied for 10 min, followed by haematoxylin counterstaining (Refine Detection Kit) for 10 min. After staining, the slides were rinsed in water, dehydrated through a graded ethanol series (70%, 90%, 100%), cleared three times in HistoClear II (National Diagnostics, HS-202) and mounted with Permount (Thermo Fisher Scientific, SP15). Image analysis was performed using Fiji software.
Catalogue numbers and dilutions of all antibodies are provided in Supplementary Table 4.
In vivo EdU/BrdU dual labelling and imaging
After sequential in vivo incorporation of EdU (20 mg per kg, intraperitoneal (i.p.), 16 h before euthanasia) and BrdU (100 mg per kg, i.p., 4 h before euthanasia), lung tissues were collected, fixed in formalin (Sigma-Aldrich, HT501128), embedded in paraffin, sectioned and mounted onto slides using standard FFPE procedures. Tissue sections were deparaffinized and rehydrated, followed by EdU staining using the Click-iT Plus EdU Cell Proliferation Kit (Invitrogen, C10637). Next, automated multiplex IF was conducted with the Leica Bond BX staining system. The sections were treated with EDTA-based epitope retrieval ER2 solution (Leica, AR9640) for 20 min at 100 °C. Primary antibodies against BrdU (Roche, 1170376), GFP (Abcam, ab13970) and Cre (BioLegend, 908001) were used. Secondary antibodies were incubated followed by nuclear counterstaining with DAPI (Sigma-Aldrich, 5 μg ml−1). The slides were mounted using Mowiol mounting reagent (Calbiochem) before imaging. Image analysis was performed using Fiji software.
In situ hybridization
mRNA in situ hybridization was performed on FFPE tissues using the manual Advanced Cell Diagnostics RNAscope 2.5 HD Reagent Kit (322350) or the RNAscope Multiplex Fluorescent Reagent Kit v2 (323100) according to the manufacturer’s instructions. Antigen retrieval times and protease digestion times were 15 and 20 min for mouse LUAD tissues, respectively. Probes for Slc4a11 and Sftpc, as well as Opal dyes from Akoya Biosciences and their dilutions for use with multiplex fluorescence in situ hybridization are listed in Supplementary Table 4.
AkaLuc in vivo bioluminescence imaging
NSG mice bearing subcutaneous transplants of KP;Slc4a11-MACD or KPfrt;Hopx-MACD reporter cells and mice bearing autochthonous KPfrt;;Hipp11GGCB/+;HopxMACD/+ LUAD tumours were i.p. injected with 100 µl of 30 mM AkaLumine-HCl substrate (Sigma-Aldrich, 808350) resuspended in PBS and imaged on the IVIS Lumina II system (PerkinElmer).
Plasma sampling and G-Luc/C-Luc measurements
Whole venous blood was collected by puncturing the submandibular vein, followed by the collection of 100 μl of blood into capillary blood collector vials (Thermo Fisher Scientific, 02-675-185). Plasma was separated by centrifugation at >8,000g for 10 min at 4 °C. For G-Luc measurement, plasma or cell culture supernatant was diluted 1:10 in PBS and 200 μM Gaussia luciferase substrate coelenterazine-h (NanoLight, 3011) was added. For C-Luc measurement, plasma or cell culture supernatant was diluted 1:100 in PBS and 0.617 μM Cypridina luciferase substrate vargulin (NanoLight, 305) was added. Luminescence was immediately measured on the BioTek Cytation 1 (Agilent) system at room temperature. This approach enables measurement of HPCS or AT1-like state growth potential (C-Luc) compared with the bulk of the tumour (G-Luc) by longitudinal measurements in the blood.
Administration of DT, MRTX1133 and cisplatin
DT (Sigma-Aldrich, D0564) was first dissolved in sterile water and diluted in sterile saline for i.p. treatment (at 50 µg per kg daily or 25 µg per kg every other day for long-term treatment and combination treatment studies). Cisplatin (West-Ward, NDC, 0143-9504-01) was dosed at 1.5 mg per kg i.p. every 3 days. MRTX1133 (a gift from Mirati Pharmaceuticals) in captisol (HY-17031, MedChem Express) at 30 mg per kg, twice per day was given i.p. as previously described9.
Lineage tracing of Slc4a11
+, Hopx
+ and random Rosa26
creERT2/+ cell state cells
Mice bearing autochthonous lung tumours were administered one or two doses of tamoxifen (200 mg per kg by oral gavage) at the indicated timepoints. For scRNA-seq analysis of Hopx+ cells and for randomly labelling Rosa26CreERT2/+ cancer cells, one dose of tamoxifen (20 mg per kg by oral gavage) was provided. The baseline measurement at 3 days was chosen to account for the conversion of tamoxifen to its active metabolite 4-OHT, recombination of the lineage-traced cells and elimination of residual 4-OHT. Tamoxifen was dissolved in corn oil at 20 mg ml−1 or 2 mg ml−1 at 60 °C for 1 h, as described previously9.
Generation of lentivirus
HEK293FreeStyle (HEK293FS) cells were transfected with lentiviral transfer plasmids and the second-generation lentiviral packaging plasmid psPAX2 (Addgene, 12260) and the envelope plasmid pMD2.G (Addgene, 12259) using either the TransIT-LT1 kit (Mirus Bio, MIR 6000) or Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, 11668500). At 24 h after transfection, the medium was discarded and replaced with fresh complete medium. Viral medium was collected and filtered through 0.45-µm PES filters (Cytiva, 6780-2504) at 48 h and 72 h after transfection. All viral media collected were concentrated using an ultracentrifuge with rotor speed set at >130,000g for 2 h at 4 °C. The supernatant was discarded into bleach and viral pellets were allowed to solubilize overnight at 4 °C. Concentrated virus was gently mixed and aliquoted. The aliquots were immediately placed on dry ice and stored at −80 °C. A fibroblast reporter GreenGo cell line expressing GFP after Flp-mediated recombination was used to titre lentivirus, as described previously66.
Generation of uPAR CAR T cells
Both mouse SFG γ-retroviral m.uPAR-m28z and human SFG γ-retroviral m.uPAR-h28z plasmids were previously described44. In the human m.uPAR-h28z CAR, the anti-mouse uPAR scFV is preceded by a human CD8a signal peptide and followed by a CD28 hinge-transmembrane-intracellular domain, a CD3z intracellular signalling domain and is linked to a P2A sequence to simultaneously express truncated LNGFR. In the mouse m.uPAR-m28z CAR, the anti-mouse uPAR scFV is preceded by a mouse CD8a signal peptide and followed by the MYC-tag sequence, mouse CD28 transmembrane and mouse CD3z intracellular domain67. Plasmids encoding the SFG γ-retroviral vectors were used to transfect gpg29 fibroblasts (H29) to generate VSV-G pseudotyped retroviral supernatants, which were used to construct stable retrovirus-producing cell lines as previously described67,68. To isolate human T cells from peripheral blood, buffy coats from anonymous healthy donors were purchased from the New York Blood Center. Peripheral blood mononuclear cells were isolated by Ficoll-based density-gradient centrifugation. T cells were purified using the human Pan T cell isolation kit (Miltenyi Biotec, 130-096-535), stimulated with CD3/CD28 T cell activator Dynabeads (Invitrogen, 11131D) as described previously69, and cultured in X-VIVO 15 (Lonza, BEBP04-744Q) supplemented with 5% human serum (Gemini Bio-Products, 100-110-100), 5 ng ml−1 interleukin-7 and 5 ng ml−1 IL-15 (PeproTech, 200-07-10UG and 200-15-10UG, respectively). T cells were counted using an automated cell counter Vi-CELL BLU (Beckman). Then, 48 h after initiating T cell activation, T cells were transduced with retroviral supernatants by centrifugation on RetroNectin-coated plates (Takara, T110B). Transduction efficiencies were determined 4 days later using flow cytometry and CAR T cells were adoptively transferred into mice or used for in vitro experiments. All blood samples were handled according to the required ethical and safety procedures. To isolate mouse T cells from peripheral blood, SV-129 and C57BL/6 mixed background mice were euthanized and the spleens collected. After tissue dissection and red blood cell lysis, primary mouse T cells were purified using the mouse Pan T cell Isolation Kit (Miltenyi Biotec, 130-095-130). Purified T cells were cultured in RPMI-1640 (Thermo Fisher Scientific, 11-875-119) supplemented with 10% FBS (GeminiBio, 900-108), 10 mM HEPES (Thermo Fisher Scientific, 15-630-080), 2 mM l-glutamine (Thermo Fisher Scientific, A2916801), MEM non-essential amino acids 1× (Thermo Fisher Scientific, 11140050), 55 µM β-mercaptoethanol (Thermo Fisher Scientific, A2916801), 1 mM sodium pyruvate (Thermo Fisher Scientific, 11360070), 100 IU ml−1 recombinant human IL-2 (Proleukin; Novartis) and mouse anti-CD3/28 Dynabeads (Gibco, 11453D) at a bead:cell ratio of 1:2. T cells were spinoculated with retroviral supernatant collected from Phoenix-ECO cells 24 h after initial T cell activation as described67,70 and used for functional analysis 3–4 days later.
Administration of uPAR CAR T cells
For uPAR CAR T transfusion into autochthonous KP tumour bearing mice, i.p. cyclophosphamide (200 mg per kg, Long Grove Pharmaceutical; NDC, 81298-8114-1) was administered to control and uPAR CAR T treatment groups. Then, 16 h later, a total of 2 × 106 mouse-derived uPAR CAR T cells per mouse was administered through i.p. injection in uPAR-CAR-T-cell-treated mice. Mice were monitored daily and collected 7 days after uPAR CAR T transfusion as indicated. For human CAR T transfusion into NSG allografts, between 5 × 106 and 7.5 × 106 human-derived CAR T cells targeting mouse uPAR were administered intratumourally per mouse.
Hyperoxia lung injury
Alveolar injury was induced in 14-week-old C57BL/6 mice by a 32 h exposure to >85% O2 in a hyperoxia chamber (BioSpherix), with FiO2 concentration maintained at a constant flow of around 3 lO2 per min and monitored by an in-line oxygen analyser. Mice were euthanized on day 7 after the 32 h exposure, followed by collection of lungs for histological analysis.
Processing of cells for droplet-based scRNA-seq
Single-cell suspensions from LUAD tumours were prepared and stained as above. The samples were multiplexed using the TotalSeq B cell hashing protocol71 (BioLegend; Supplementary Table 4). Live sorted cells were collected by flow cytometry, washed once with PBS containing 1% BSA and resuspended to a final concentration of 700–1,300 cells per μl of PBS + 1% BSA and processed by droplet-based scRNA-seq as below.
scRNA-seq
Single-cell suspensions were stained with Trypan blue, and the Countess II Automated Cell Counter (Thermo Fisher Scientific) was used to assess both the cell number and viability. After quality control, the samples were loaded onto Next GEM Chip G (14143, 15123, 15342, 15488, 15600, 15601, 15771) or GEM-X Single Cell Chip (16235, 16318, 16562, 16686, 17402, 17483, 17543, 17721) (10x Genomics PN 1000690 and 2000060) and GEM generation, cDNA synthesis, cDNA amplification and library preparation of around 40,000–50,000 cells proceeded using the Chromium Next GEM Single Cell 3′ Kit v3.1 or GEM-X Single Cell 3′ Kit v4 (10x Genomics, 1000268 and 1000691) according to the manufacturer’s protocol. cDNA amplification included 11–12 cycles, and 78–863 ng of the material was used to prepare sequencing libraries with 8–14 cycles of PCR. Indexed libraries were pooled and sequenced on the NovaSeq 6000 (14143, 16235, 16562) or X (15123, 15342, 15488, 15600, 15601, 15771, 16235, 16318, 16562, 16686, 17402, 17483, 17543, 17721) system in a PE28/88 run using the NovaSeq 6000 S4 (200 Cycles) or X 10B (100 Cycles) or 25B (300 cycles) Reagent Kit (Illumina). An average of 38,000 paired reads was generated per cell.
Cell surface protein feature barcode analysis
Amplification products generated using the methods described above included both cDNA and feature barcodes tagged with cell barcodes and unique molecular identifiers. Smaller feature barcode fragments were separated from longer amplified cDNA using a 0.6× cleanup using aMPure XP beads (Beckman Coulter, A63882). Libraries were constructed using the 3′ Feature Barcode Kit (10x Genomics, 1000276) according to the manufacturer’s protocol with 10–12 cycles of PCR. Indexed libraries were pooled and sequenced on the NovaSeq 6000 (14143_B,16562_B) or X (15123_B, 15342_B, 15488_B, 15600_B, 15601_B, 15771_B, 16235_B, 16318_B, 16562_B, 16686_B, 17402_B, 17543_B) system in a PE28/88 run using the NovaSeq 6000 S4 (200 Cycles) or X 10B (100 cycles) or 25B (300 cycles) Reagent Kit (Illumina). An average of 359 million paired reads was generated per sample.
Computational analyses
Jupyter notebooks executing the analysis workflow and figure generation are available on GitHub (https://github.com/dbetel/HPCS_LUAD). All generated sequencing data and count matrices are available at the NCBI Gene Expression Omnibus under accession number GSE277777.
Processing of scRNA-seq data
FASTQ files of scRNA-seq data generated on the 10x Chromium or ChromiumX platform were processed using the standard CellRanger pipeline (version ≥6.2)72. Reads were aligned to a custom GRCm38/mm10 reference genomes including the eGFP, TagBFP, tdTomato, Gluc, Cluc, Akaluc, mScarlet, creERT2 and DTR transgenes. Cell-gene count matrices were analysed using a combination of published packages and custom scripts centred around the SCANPY/AnnData ecosystem73. scRNA-seq datasets from different mouse models and primary patient samples were analysed separately using similar workflows.
scRNA-seq data were compiled into a combined count matrix. In general, cells with less than 300 unique molecular identifiers (UMIs), more than 10–20% mitochondrial UMIs and low complexity based on the number of detected genes versus number of UMIs were removed as indicated in the available code. Where applicable, doublets were filtered by modelling the TotalSeq B hash count distribution as a Bayesian Gaussian mixture model with variational inference74. The same method was used to demultiplex the sample into individual hashes. UMI counts were normalized using the default CPM normalization. In the case of non-hashed transplant samples, the R package scDblFinder75 was used to detect doublets, which were then removed before further analysis.
To identify highly variable features, variance-stabilizing transformation and dimensionality reduction were performed on normalized, log2-transformed count data using principal component analysis. The resulting dimensionality-reduced count matrices were used as an input for uniform manifold approximation and projection embedding and unsupervised clustering using the Leiden algorithm76.
Cell state classifications
To compare data generated from the droplet based 10x Chromium platforms with our previous work4, we first identified the common genes between each new 10x dataset and our previously published single-cell dataset, which was generated using the SmartSeq2 method77. For each 10x dataset, we trained a multiclass logistic regression model using the scikit-learn LogisticRegression class with options multi_class=’multinomial’ and solver=‘lbfgs’ using our original cluster labels (hereafter, cell state identities) and gene counts from our previous work4, using only the genes in common between both datasets. We then used this model to classify the cells generated in our 10x dataset. To assign cell state identities to each Leiden cluster generated from our SCANPY pipeline above, we took a pluralistic voting approach in which the cell states that were the most represented in a Leiden cluster were used as that cluster’s cell state identity. A rare exception to this were those Leiden clusters where a significant proportion of cells were identified as highly proliferative and were subsequently shown to have high Mki67 expression. These Leiden clusters were assigned a highly proliferative cell state identity. Cell state assignments can be found in the source code available at the GitHub repository above. Notably, based on recent work identifying a hybrid lung/gastric-like cell state expressing Nkx2-1 and Hnf4α40,41, we reannotated cells classified as clusters 8 and 10 in our original work4 to comprise the hybrid lung/gastric-like cell state.
Marker evaluation for classification of cell states
The presence of marker transcripts was determined on processed cell counts as transcript levels greater than the minimum value detected by scRNA-seq (that is, non-zero counts). Cell state classifications were determined as above, with the modification that putative transcript markers were removed as input factors from all processing that could bias cell state scores and assignments towards the HPCS. We calculated true-positive (TP), false-positive (FP), true-negative (TN) and false-negative (FN) metrics and calculated marker sensitivity, specificity, positive predictive values, and negative predictive value as below:
$$\mathrm{Sensitivity}=\frac{\mathrm{TP}}{\mathrm{TP}+\mathrm{FN}}$$
$$\mathrm{Specificity}=\frac{\mathrm{TN}}{\mathrm{TN}+\mathrm{FP}}\,$$
$${\rm{P}}{\rm{o}}{\rm{s}}{\rm{i}}{\rm{t}}{\rm{i}}{\rm{v}}{\rm{e}}\,{\rm{p}}{\rm{r}}{\rm{e}}{\rm{d}}{\rm{i}}{\rm{c}}{\rm{t}}{\rm{i}}{\rm{v}}{\rm{e}}\,{\rm{v}}{\rm{a}}{\rm{l}}{\rm{u}}{\rm{e}}=\frac{{\rm{T}}{\rm{P}}}{{\rm{T}}{\rm{P}}+{\rm{F}}{\rm{P}}}$$
$${\rm{N}}{\rm{e}}{\rm{g}}{\rm{a}}{\rm{t}}{\rm{i}}{\rm{v}}{\rm{e}}\,{\rm{p}}{\rm{r}}{\rm{e}}{\rm{d}}{\rm{i}}{\rm{c}}{\rm{t}}{\rm{i}}{\rm{v}}{\rm{e}}\,{\rm{v}}{\rm{a}}{\rm{l}}{\rm{u}}{\rm{e}}=\frac{{\rm{T}}{\rm{N}}}{{\rm{T}}{\rm{N}}+{\rm{F}}{\rm{N}}}$$
For similar calculations using the SmartSeq2 LUAD data4 from which the signatures were originally derived, we reprocessed raw count data using the pipeline described above up to the Leiden clustering step. We then calculated gene scores using the built-in SCANPY score_genes function using the top 100 genes from the previously determined HPCS gene signature4 (Supplementary Table 5), again with the marker genes removed to prevent bias. We labelled clusters enriched for the HPCS gene signature as HPCS clusters and calculated the sensitivity, specificity, positive predictive value and negative predictive values as described above for the marker gene of interest. The results for all sensitivity, specificity, positive predictive value and negative predictive values for marker genes are provided in Supplementary Tables 1–3. We estimate the sensitivity of the HPCS to be 9.26% and 65.4% based on the 10x droplet-based and SmartSeq2 scRNA-seq data, respectively, while the specificity is 99.7% and 84.3%. Although we acknowledge that we cannot completely rule out the possibility that a minor proportion of non-HPCS cells are traced in our experiments, based on the specificity of the reporter system as calculated above, this is likely to be an insignificant minority of cells.
Phenotypic volume calculations
Phenotypic volumes, quantitative measures capturing the diversity of cellular phenotypes in cell populations, were performed on highly variable genes, as previously described37. Distributions of phenotypic volumes were calculated by sampling 100 cells randomly with replacement from the cells of interest and calculating the phenotypic volume for 1,000 replicates per cell population. Statistical significance was determined using either t-tests or ANOVA to compare the distributions of phenotypic volumes.
Gene signature score calculation and correlation
Gene signatures were compiled from a variety of sources listed in Supplementary Table 5 and were used to calculate scores using the SCANPY score_genes function. Scatter plots and Pearson correlations were generated by independently calculating each cell’s gene signature score and HPCS score and then comparing the distribution of scores. Lines of best fit and r2 values were calculated using scipy.stats.linregress and statistical significance for Pearson correlations was determined using an exact distribution with the built-in scipy.stats.pearsonr function. Gene signature scores were used to compare cells with or without Slc4a11 expression as indicated.
External data analysis
All gene signatures and analysed datasets from previously published works are listed in Supplementary Table 5.
Time-series analysis
SmartSeq2 scRNA-seq LUAD data (GEO: GSE152607)4 were downloaded from the NCBI GEO. scRNA-seq data from wild-type AT2 cells, KP adenoma and KP adenocarcinoma tumour data from the 2, 12, 20 and 30 week post-tumour induction timepoints were used with Moscot’s78 TemporalProblem to determine intertimepoint couplings through optimal transport, and the RealTimeKernel.from_moscot() function was used to convert the intertimepoint couplings into a CellRank79 transition matrix. This matrix was used with a generalized perron-cluster cluster analysis estimator to identify terminal macrostates. Gene expression trajectories to the terminal macrostates were plotted with the Cellrank gene_trends function using a generalized additive model and the time component was determined by Palantir80 pseudotime. The relative estimated start and end of the HPCS was modelled in Palantir pseudotime by plotting the trend of a calculated HPCS score and comparing it to known marker genes using the above process. The calculated HPCS score was defined using the top 100 genes of the HPCS gene signature (Supplementary Table 5) with the SCANPY score_genes function.
Software versions
SCANPY (v. ≥1.9), pingouin (v.0.5.4), gseapy (v.1.1.1), numpy (v. ≥1.26), scipy (v. ≥1.12), scikit-learn (v. ≥1.13), leidenalg (v.0.10.2), matplotlib (v.3.8.4), Cellrank (v.2.0.7), Palantir (v.1.4.1), R (v.4.3.3), FIJI/ImageJ (v. >1.54) and GraphPad (v. >9.0) were used.
Statistics and reproducibility
Statistical analyses were performed using Student’s t-tests, Welch’s tests, Mann–Whitney U-tests, Kruskal–Wallis tests, Holm–Šídák tests, one-way ANOVA or two-way ANOVA, as appropriate. All statistical analyses performed were in a two-sided manner. Statistical significance for the figures is indicated as raw P values or with asterisks. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant. Representative micrographs from experiments were performed at least three times independently with similar results. For Fig. 5f, hyperoxia injury was performed at three different timepoints for one mouse each, with Slc4a11-related expression peaking at 7 days. All of the experiments were performed as biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.