Mice
WT C57BL/6 and CD45.1 SJL mice, mice expressing Cre recombinase (LysM–Cre) under the control of the lysozyme (LysM) promoter, CX3CR1–ERT2 inducible Cre, Rosa26-eYFP (R26YFP) mice and RiboTag mice61 harbouring a modified allele of Rpl22 (Rpl22-HA) induced by the action of Cre recombinase were all purchased from Jackson Laboratories. Dhps floxed mice were a gift from S. Balabanov, Zurich. Mice were bred and maintained under specific-pathogen-free conditions, and experiments were performed under protocols approved by the Regierungspräsidium Freiburg, the Animal Welfare Committee of the Max Planck Institute of Immunobiology and Epigenetics, or the Institutional Animal Care and Use Committee of Johns Hopkins University School of Medicine, in accordance with the Guide for the Care and Use of Animals. Mice used for all experiments were littermates and matched for age and sex; both male and female mice were used. Mice of all strains were typically 6–12 weeks of age unless otherwise specified. Experiments were not performed blinded. Parabiosis experiments were performed in the laboratory of F.G., with Institutional Animal Care and Use Committee approval from the Biological Resource Center (A*STAR) in Singapore, as follows: the animals were monitored daily for evidence of fighting and were exposed to a gel food diet supplement at least two times, 2–6 weeks before performance of parabiotic attachment. Animals were randomly assigned a pair member, with the appropriate transgene or CD45 congenic allotype, within their cohoused cohort. Pairs established for parabiotic attachment were ‘cage paired’, meaning they were housed only with their future parabiotic pair member, 2 weeks before the parabiosis procedure, and the mice were observed again for aggressive behaviour. Aggressive and injured mice were removed from the study. With this husbandry protocol, aggression was highly uncommon.
Tissue processing for RTM isolation
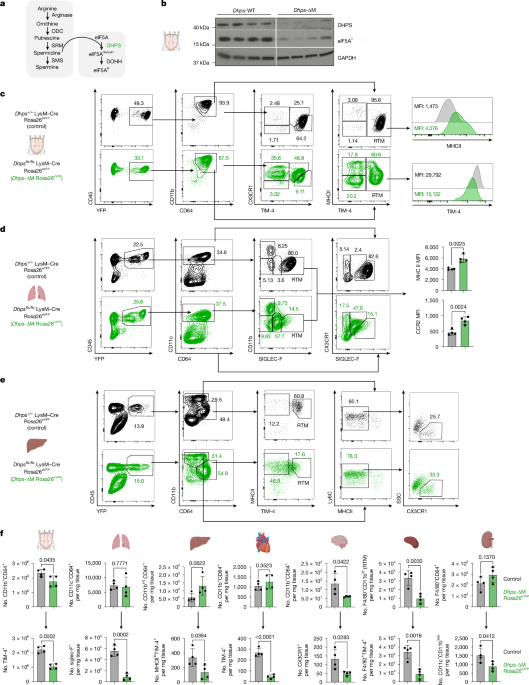
Lungs, liver, kidney, brain and heart were harvested after perfusion with ice-cold phosphate-buffered saline (PBS) through the heart; organs were placed in 5-ml tubes containing 2 ml digestion solution with 2 mg ml−1 collagenase IV (Thermo Fisher) and 0.1 mg ml−1 DNAse I (Sigma) and cut into small pieces with scissors. The solution was incubated in a shaker at 37 °C for 1 h. Tissues were further homogenized using a 20 G × 1” needle and a 5-ml syringe, and homogenates were filtered through a 100-μm cell strainer. RBCs from lungs, liver, kidney and heart were lysed with ACK for 2 min, washed with PBS 1% fetal calf serum (FCS), 2 mM EDTA (FACS buffer) and kept on ice until flow cytometry staining. For splenic macrophages, no digestion solution was used. Cells were isolated by mechanical disaggregation of spleen on a 70-μm strainer, RBCs were lysed and cells were resuspended in FACS buffer. Microglia were isolated postdigestion using a 35–70% Percoll gradient with centrifugation for 20 min at 500g and room temperature without brakes. The leucocyte layer was transferred to tubes containing FACS buffer and kept on ice until flow cytometry staining. When indicated, alveolar macrophages were isolated by BAL and centrifuged to isolate lavage fluid for optical density measurement, and the pelleted cells were resuspended in FACS buffer for flow cytometry staining. Absolute numbers of macrophages were calculated using 123count eBeads Counting Beads (Thermo Fisher) following the manufacturer’s instructions. For solid organs, the weight of each tissue in milligrams was recorded after harvesting, and the number of macrophages per sample was normalized by this value to obtain the total cells per milligram tissue.
Peritoneal macrophages in vitro culture
Peritoneal macrophages were obtained by peritoneal lavage and either cultured in complete medium (RPMI 1640 medium supplemented with 10% FCS, 2 mM L-glutamine and 100 U ml−1 penicillin/streptomycin) with 20 ng ml−1 macrophage colony stimulating factor (PeproTech) on a six-well-plate, or transferred to a 96-well-plate and used directly for flow cytometry staining. For in vitro activation, peritoneal macrophages were cultured with 20 ng ml−1 IL-4 (PeproTech) overnight to generate alternatively activated macrophages. For IL-33 (R&D Systems) in vitro treatment, 20 ng ml−1 was used in all experiments unless otherwise specified, with culture for 72 h.
Parabiosis
Parabiotic mice were generated as reported62 from age- and weight-matched CD45.2+ (Dhpsflx/flx LysM–Cre negative (Dhps-WT) or Dhpsflx/flx LysM–Cre positive (Dhps-ΔM) and CD45.1+ (C57BL/6) mice (WT) between 6 and 8 weeks old.
Bone marrow chimera
CD45.1+ (C57BL/6) mice (WT) were irradiated (1,200 cGy, 2 split doses, 3 h apart) in a Cesium Mark 1 irradiator (JL Shepperd & Associates) and then infused with a mix of 5 × 106 bone marrow cells from Dhpsflx/flx LysM–Cre negative (Dhps-WT) or Dhpsflx/flx LysM–Cre positive (Dhps-ΔM) CD45.2+ cells mixed with 5 × 106 bone marrow cells from CD45.1+ mice (1:1 ratio). Mice were harvested, and the frequency of CD45.1+/CD45.2+ RTMs were analysed at 12 weeks after bone marrow transfer.
In vivo proliferation of peritoneal macrophages by IL-4c administration
For long-acting IL-4 treatment, a mixture of 5 mg recombinant mouse IL-4 (Peprotech) and 25 mg anti-IL-4 mAb (clone 11B11; BioXcell) was incubated for 5 min on ice to form an IL-4:anti-IL-4 complex (IL-4c). IL-4c enables sustained and slow IL-4 release. Mice were then injected i.p. with 100ul of IL-4c (containing 5 μg IL-4 and 25 μg anti-IL-4), or PBS vehicle control. The peritoneal lavage was analysed at day 4 post-injection. To measure proliferation, EdU (0.5 mg) was injected i.p. at day 0 and day 2 in IL-4c treated mice and analysed at day 4. To assess proliferation of adoptively transferred peritoneal macrophages, ex vivo isolated peritoneal macrophages from Dhps-WT or Dhps-ΔM CD45.2 mice were stained with CTV (Life Technologies) following the manufacturer’s instructions and assessed for purity by flow cytometry; then, 0.5 × 106 to 1.0 × 106 cells per mouse were transferred by injection i.p. to CD45.1 WT receptor mice. Immediately after transfer, IL-4c or PBS was injected i.p., and macrophage proliferation was measured at day 4 by CTV dilution with flow cytometry.
RTM in vivo depletion by CL treatment
Mice were injected with 200 μl CL (Liposoma BV) i.p. or PBS to deplete RTMs in the peritoneal cavity and liver. Peritoneal lavage and tissues were collected at the indicated time points for flow cytometry or tissue histology.
For depletion of alveolar macrophages, adult male or female mice were treated with intratracheal delivery of CL at days −2 and 0 after the following procedure on each day: mice were anaesthetized with a mixture of ketamine (75 mg kg−1) and xylazine (10 mg kg−1) injected i.p. Their necks were cleaned with alternating scrubs of 70% ethanol and betadine. A vertical incision was then made in the neck, and the trachea was exposed. Orotracheal intubation was performed using a 20 g intravenous catheter under direct visualization of the trachea. The mice were briefly ventilated using a MiniVent Model 845 (Harvard Apparatus) with a respiratory rate of 150 breaths per min and tidal volume of 200 μl. The mice were then disconnected from the ventilator, and 70 μl of liposomes were delivered each day. The mice were again briefly ventilated and then extubated and allowed to breathe spontaneously. The neck incision was closed with glue. The mice were kept on a warming blanket until they awakened from anaesthesia and then returned to their cages. BAL was collected at different time points posteuthanasia for flow cytometry analysis. Control mice received PBS instead of CL.
In vivo Cre induction by tamoxifen administration
Tamoxifen (Sigma) was dissolved in corn oil to a final concentration of 10 mg ml−1 and stored at −20 °C. Adult Cx3cr1CreERT2-Rosa26YFP mice were given 200 μl (2 mg) tamoxifen solution once per day for 4 consecutive days, injected i.p. Tissues were harvested at the indicated time points after tamoxifen injection.
In vitro efferocytosis of apoptotic cells by ex vivo peritoneal macrophages
Peritoneal macrophages were plated on 24-well uncoated tissue culture plates at a density of 0.1 × 106 cells per well overnight before addition of apoptotic cells. Apoptotic cells were prepared from total lymphocytes isolated from lymph nodes and treated with staurosporin at 0.5 μM overnight at a density of 5 × 106 ml−1 to induce apoptosis. Apoptotic cells were then washed with culture medium and stained with CypHer5E NHS Ester (Cytiva) and CTV (Thermo Fisher), each at 1 μM in PBS. Labelled apoptotic cells were washed three times in culture medium (containing 10% FBS), pelleted and resuspended for counting by trypan blue exclusion.
Apoptosis was confirmed by Apotracker and live/dead viability staining with flow cytometry. Apoptotic cells were added to macrophages in a 1:5 ratio (macrophages to apoptotic cells). To increase the contact between macrophages and apoptotic cells, 30 s centrifugation at 300g was performed, and cells were left at 37 °C in an incubator for 30 min. After this time, apoptotic cells were washed three times with culture medium, and macrophages were left for another 60 min at 37 °C to allow progression of efferocytosis. To analyse the rate of efferocytosis, macrophages were collected and stained for flow cytometry. Cargo-positive cells were gated as F4/80+CTV+, and levels of efferocytosis were analysed on the basis of MFI of cypher5e in the AF647/APC channel in cargo-positive cells.
Mouse sRBC preparation for in vitro and in vivo transfer
Fresh RBCs were prepared by isolation from mouse peripheral blood and depletion of leukocytes by two steps. First, a 35–70% Percoll gradient was used to pellet RBCs and deplete leukocytes (positioned in the interphase). The second step involved incubating the RBC pellet with mouse CD45 MojoSort beads (BioLegend) following the manufacturer’s protocol to deplete any remaining contaminating CD45+ cells. After isolation, RBCs were stressed by heat shock at 48 °C for 20 min under constant agitation. The generated sRBCs were spun down for 10 min at 400g in PBS to remove free haemoglobin and colabelled with CTV and Cypher5e following the manufacturer’s instructions. Before use, sRBCs were washed more than three times with cold PBS to remove free CTV and Cypher5e probes.
In vitro efferocytosis of sRBCs by ex vivo peritoneal macrophages
Peritoneal macrophages were plated on 24-well uncoated tissue culture plates at a density of 0.1 × 106 cells per well overnight before addition of sRBC. To increase the contact between macrophages and sRBCs, a 30-s centrifugation at 300g was performed, and cells were left at 37 °C in an incubator for 30 min. After this time, sRBCs were washed three times with culture media, and macrophages were left for another 60 min at 37 °C to allow progression of efferocytosis. For analysis of the rate of efferocytosis, macrophages were collected and stained for flow cytometry. The percentage of macrophages in active efferocytosis was gated as CTV+Cypher5+ in the F4/80+CD11b+ gate, and the progression of efferocytosis was analysed by MFI of cypher5e in the AF647/APC channel within this gate.
In vivo removal of transferred stressed sRBCs from peripheral blood
CTV and Cypher5e colabelled sRBCs were prepared as described above and injected into mice through the tail vein. The prepared sRBCs (isolated from one mouse) were injected into ten recipient mice. Each recipient mouse received a total volume of sRBCs resuspended in 200 μl of PBS. At 10 min (initial sRBC frequency in circulation) and 90 min (end time point) after injection, mice were euthanized, and whole blood was isolated. The frequency of remaining labelled sRBCs was measured by flow cytometry by gating in CD45−CTV+Cypher5e+ cells in peripheral blood.
BAL and ex vivo lung proteinosis analysis
A vein catheter (BD) was inserted into the trachea of mice, and the first wash was performed with 1 ml of prewarmed PBS followed by four extra washes to collect BAL cells. The first BAL wash with PBS was centrifuged for 5 min at 500g, and the supernatant was used for measurement of optical density at 600 nm using a spectral photometer. The cell pellet was merged with the rest of the BAL washes for analysis by flow cytometry.
Flow cytometry protocol and cell sorting
For flow cytometry staining, cells were washed in cold PBS, and non-specific antibody binding to cells was blocked by incubation with an anti-CD16/32 antibody (clone 2G8; BD Biosciences) at 4 °C for 10 min in the presence of LIVE/DEAD viability dye (Thermo). Then, cells were stained with fluorophore-conjugated antibodies (Supplementary Table 9) at 4 °C for 25 min in PBS 1% FCS, 2 mM EDTA (FACS buffer). Cells were maintained at 4 °C and analysed on a BD Fortessa X20, Celesta or Symphony (BD Biosciences). For intracellular staining of active caspase-3 and Ki-67 (Supplementary Table 9), BD Cytofix/Cytoperm Fixation/Permeabilization Kit was used following manufacturer recommendations and the cells stained overnight and analysed on a BD Fortessa X20, Celesta or Symphony (BD Biosciences). Data were analysed in FlowJo (FlowJo LLC). For cell sorting non-specific antibody binding to cells was blocked by incubating cells with an anti-CD16/32 antibody (clone 2.4G2; BD Biosciences) at 4 °C for 10 min. The cells were then stained with fluorophore-conjugated antibodies (Supplementary Table 9) at 4 °C for 25 min. FACS was performed on a BD FACS Aria III (BD Biosciences) to achieve greater than 95% purity. Dead cells were excluded by staining with LIVE/DEAD viability dye (Thermo).
Western blot protocol
For western blot analysis, cells were washed with ice-cold PBS and lysed in 1× Cell Signaling lysis buffer (20 mM Tris-HCl, (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg ml−1 leupeptin (Cell Signaling Technologies), supplemented with 1 mM phenylmethylsulfonyl fluoride. Samples were frozen and thawed 3 times, followed by centrifugation at 20,000g for 10 min at 4 °C. Cleared protein lysate was denatured with LDS loading buffer for 10 min at 70 °C and loaded on precast 4% to 12% Bis-Tris protein gels (Life Technologies). Proteins were transferred on to nitrocellulose membranes using an iBLOT 2 system (Life Technologies) following the manufacturer’s protocols. Membranes were blocked with 5% w/v milk and 0.1% Tween-20 in Tris-buffered saline (TBS) and incubated with the appropriate antibodies in 5% w/v bovine serum albumin in TBS with 0.1% Tween-20 overnight at 4 °C. All primary antibody incubations were followed by incubation with secondary HRP-conjugated antibody (Pierce) in 5% milk and 0.1% Tween-20 in TBS and visualized using SuperSignal West Pico or Femto Chemiluminescent Substrate (Pierce) on Biomax MR film (Kodak) or a Bio-Rad ChemiDoc. The following antibodies were used: anti-DHPS (Abcam), anti-GAPDH, anti-EIF5A (BD Bioscience), anti-EIF5A-hypusine (Millipore). All antibodies were used at a dilution of 1:1,000.
Mouse tissue histology and histopathologic staining
Perfused lungs, liver, kidney, heart, spleen and brain were fixed overnight in 10% formalin at 4 °C. Lungs were inflated with 1 ml of formalin before harvesting. All organs were cut into 4-μm-thick formalin-fixed paraffin-embedded (FFPE) tissue sections. Liver sections were stained with H&E or MAS. All sectioning and staining were performed by The Johns Hopkins University Oncology Tissue and Imaging Services Core Laboratory. H&E- and MAS-stained liver slides from CL-treated mice and control images were observed and pathologically described by Y.Z. Immunohistochemical staining was performed by the Johns Hopkins University Oncology Tissue and Imaging Services Core Laboratory. Lung sections were stained with an antibody against F4/80 (brown) and lung tissues using antibodies against CD31 for vasculature (purple), EpCAM for epithelial cells (teal) and haematoxylin (indigo-like blue) for nuclei. H&E, MAS and immunohistochemically stained slides were whole-slide scanned using Hamamatsu NanoZoomer XR and NDP software (Hamamatsu Photonics, NDP.scan). NDP.view2 (Hamamatsu Photonics) was used for image analysis.
Immunofluorescence staining and image acquisition of FFPE tissue sections
For immunofluorescence staining of lung, liver, kidney, heart, spleen and brain with FFPE sections of mouse tissues, heat-induced epitope retrieval with Diva Decloaker (Biocare Medical, DV2004MX) was performed after deparaffinization and rehydration. Then, slides were blocked with 2.5% normal donkey serum (Jackson ImmunoResearch, 017-000-121) for 30 min at room temperature and incubated with the cocktail of primary antibodies for 16 h at 4 °C. Slides were washed with PBST (Dulbecco’s phosphate-buffered saline with 0.05% Tween-20), then incubated with the cocktail of secondary antibodies conjugated with fluorochrome for 2 h at room temperature. A Streptavidin/Biotin Blocking Kit (VectorLabs, SP-2002) was used before the incubation with antibodies when endogenous biotin blocking was needed. Slides were cover-slipped with EverBrite TrueBlack Hardset Mounting Medium with DAPI (Biotium, 23018) or ProLong Gold Antifade Mountant (Invitrogen, P36930) after nuclear staining with 1 μg ml−1 DAPI (Thermo Scientific, 62248) for 10 min at room temperature. Images were acquired with Yokogawa Spinning Disk Field Scanning Confocal Systems (Nikon, CSU-W1 SoRa), ZEISS Axioscan 7 or Vectra Polaris (Akoya Biosciences). Images were analysed using NIS Elements (Nikon), ImageJ (v.1.54f, National Institutes of Health), ZEN (ZEN lite, v.3.9.101, ZEISS) and QuPath-0.5.0-x64. For lung sections, five image fields per sample were selected to be analysed.
For the measurement of macrophages of lung (cell counts, area and circularity), five image fields (202,536 μm2 for each field) per sample were randomly selected from whole-slide scanned images using ZEISS Axioscan 7 and ZEN software. Random fields were at least 800 μm away from each other; this was confirmed after the selection. The formula (circularity) = 4π × (area)/(perimeter)2 was used to calculate sphericity, with a value close to 1 indicating a round object (sphericity = 1 for an exact circle). ImageJ was used for measurement. The following primary antibodies were used: anti-PDGFRα (R&D Systems, AF1062, polyclonal, 1:200 dilution), anti-F4/80 (Bio-Rad, MCA497GA or MCA497B, clone Cl:A3-1, 1:300 or 1:50), anti-CD64 (Invitrogen, MA5-29706, clone 027, 1:300), anti-GFP (Abcam, for YFP detection, ab5450, polyclonal, 1:400), anti-IBA1 (Synaptic Systems, HS-234 308, clone Gp311H9. 1:300) and anti-TIM-4 (BioLegend, 130002, clone RMT4-54, 1:400). Streptavidin conjugated with Alexa Fluor 488 (Invitrogen, S32354) or Alexa Fluor 594 (Invitrogen, S32356) was used for biotin-conjugated primary antibody detection. The following secondary antibodies conjugated with fluorochrome were used: Alexa Fluor Plus 488 (Invitrogen, A32814 and A32790), Alexa Fluor Plus 555 (Invitrogen, A48270 and A32794), Alexa Fluor 594 (Jackson ImmunoResearch, 706-585-148) and Alexa Fluor Plus 647 (Invitrogen, A32795 and A32849).
Immunofluorescence staining of frozen sections from mouse kidney and brain
After transcardial perfusion with PBS, kidneys and brains were harvested and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, catalogue no. 15710) at 4 °C for at least 24 h. Fixed tissues were dehydrated in 30% sucrose solution (Sigma, S1888) for at least 48 h and embedded in Tissue-Plus O.C.T. compound (Fisher Scientific, 23-730-571). Cryosections, prepared by The Johns Hopkins University Oncology Tissue and Imaging Services Core Laboratory, were obtained at a thickness of 10 μm (kidney) or 15 μm (brain). Cryosections were air-dried and blocked for 1 h with 2.5% normal donkey serum (Jackson ImmunoResearch, 017-000-121) and 0.3% Triton X-100 (Sigma) in PBS, followed by incubation with anti-GFP (Abcam, ab5450, 1:500), anti-IBA1 (Invitrogen, PA5-27436, 1:500) diluted in blocking buffer at 4 °C overnight. For anti-cleaved caspase-3 (CST, 9661, polyclonal, 1:300) staining, epitope retrieval was performed using Antigen Retrieval Citra Plus (Biogenex, HK080-9K). After washing, sections were incubated with secondary antibodies (1:500) conjugated with Alexa Fluor Plus 647 (Invitrogen, A32849) and Alexa Fluor 568 (Invitrogen, A10042,) at room temperature for 1 h. Slides were cover-slipped with mounting medium (Vector Laboratories, H-1900-10), after staining of nuclei with 1 μg ml−1 DAPI (Sigma, D8417) for 3 min at room temperature. Images were captured using a confocal microscope (Zeiss LSM880 Airyscan). For kidney, at least four fields per sample were randomly captured and analysed using Imaris software (10.0.1, Bitplane) to measure the volume and sphericity of YFP-expressing macrophages. For cleaved caspase-3 analysis, whole-slide scanning was performed with a Vectra Polaris (Akoya Biosciences), and images were analysed using QuPath-0.5.0-x64. For brain, images of the hippocampal region were captured and denoised using ImageJ (v.1.54g, National Institutes of Health). The volume and sphericity of microglia were analysed using Imaris (10.0.1, Bitplane).
Proteomics procedure and analysis
Protein samples were prepared with 5 × 106 cells using an iST 96X kit (PreOmics), according to the manufacturer’s recommendations. All samples used for data-dependent acquisition (DDA) and data-independent acquisition (DIA) analyses were spiked with index retention time kit peptides (Biognosys), according to the manufacturer’s instructions. DIA spectral libraries were generated with Spectronaut (Biognosys) v.10.0 using MaxQuant (https://www.maxquant.org/) results as an input63. For the latter, DDA runs (using two or three biological replicates from each biological conditions) were acquired using a Q Exactive Plus instrument, and data were searched using MaxQuant (v.1.6.1.0). The spectral library was constructed using a false discovery rate cutoff of 1% and minimum and maximum of 3 and 6 fragment ions, respectively, and protein grouping was performed according to the MaxQuant search results.
For mass spectrometric acquisition, the general nanoscale liquid chromatography mass spectrometry setup was similar to that previously described63 with minor modifications. A Q Exactive Plus mass spectrometer (Thermo Fisher) and an Easy nanoLC-1200 (Thermo Fisher) were used for both DDA and DIA experiments. For the chromatographic separation of peptides, 4 μg peptide digest was analysed at 50 °C (controlled by a Sonation column oven) on a 50-cm in-house-packed fused-silica emitter microcolumn (75 μm inner diameter, 360 μm outer diameter, 8 µm tapered open end; SilicaTip PicoTip; New Objective) packed with 1.9-μm reverse-phase ReproSilPur C18-AQ (120 Å) beads (Dr Maisch). Peptides were separated using a binary solvent system consisting of 0.1% (v/v) formic acid (solvent A) and 80% (v/v) acetonitrile/0.1% (v/v) formic acid (solvent B). Peptides were loaded at 400 nl min−1 at 0% B and then separated by the following linear gradients: from 2% to 10% solvent B over 5 min at 250 nl min−1, from 10% to 35% over 180 min at 250 nl min−1, from 35% to 50% over 26 min at 250 nl min−1, and from 50% to 80% over 6 min at 250 nl min−1, and kept at 80% for 10 min at 250 nl min−1, followed by an inverse gradient from 80% to 5% over 6 min and a re-equilibration step over 5 min at 5% B (300 nl min−1).
For data analysis, MS2-based label-free quantification was carried out by processing DIA raw data using Spectronaut (v.10.0) software with default parameters as previously described63 with minor modifications. In brief, the decoy method was set to ‘mutated’, data extraction and extraction window were set to ‘dynamic’ with correction factor 1, identification was set to ‘normal distribution P-value estimator’ with q-value cutoff of 0.1 and the profiling strategy was set to ‘iRT profiling’ with q-value cutoff of 0.01. Ultimately, protein quantity was set to ‘average precursor quantity’ and the smallest quantitative unit was set to ‘precursor ion’ (summed fragment ions). For statistical testing and identification of deregulated proteins in all approaches, a two-sample Student’s t-test was used to identify differentially expressed proteins filtered to 1% false discovery rate.
scRNA-seq procedure and analysis
For peritoneal macrophage sequencing, single cells were isolated from the peritoneal cavities of the indicated animals with 3–4 biological replicates per genotype (Dhps-WT and Dhps-ΔM) and prepared for scRNA-seq using the GemCode Single Cell Platform with GemCode Gel Beads, Chip and Library Kits (v.3) and 10x Genomics Chromium Controller following the manufacturer’s protocol. Peritoneal lavage libraries were sequenced on a NovaSeq 6000 (Illumina). For lung macrophage sequencing, single cells were isolated from digested lungs as described before, and LIVE CD45+YFP+ cells were FACS sorted from three biological replicates per genotype for Dhps+/+ LysM–Cre Rosa26eYFP (control) and Dhps-ΔM Rosa26eYFP mice. Cells were submitted to sequencing company OMAPiX for 10x Genomics library preparation using the GEM-X 3′ v.4 gene expression kit. Lung libraries were sequenced on a NovaSeq X Plus (Illumina).
Samples were demultiplexed, quality checked, filtered and aligned with genome build GRCm38 using pre-established pipelines implemented in snakePipes64 with STARsolo v.2.7.4a65, deeptools v.3.3.2, seqtk v.1.3, pigz v.2.3.4, snpsplit v.0.3.4, samtools v.1.10, fastqc v.0.11.9, cutadapt v.2.8, trim-galore v.0.6.5, multiqc v.1.8, fastp v.0.20.0, umi_tools v.1.0.1 and star v.2.7.4a. For lung macrophage libraries, CellRanger v.9.0.1 was used to obtain count matrices. Resulting raw read count matrices of barcodes corresponding to cells and features corresponding to detected genes were processed, analysed and visualized in R v.4.3.1 (ref. 66) using Seurat v.4 (ref. 67) with default parameters for all functions unless otherwise specified. Poor-quality cells with low total unique molecular identifier counts and high percentages of mitochondrial gene expression were excluded. Filtered samples were normalized using a regularized negative binomial regression (SCTransform)67 and integrated with the reciprocal principal component analysis approach followed by mutual nearest neighbours, using 50 principal components. Integrated gene expression matrices were visualized with UMAP68 as a dimensionality reduction approach. Resolution for cell clustering was determined by evaluating hierarchical clustering trees at a range of resolutions (0–1.2) with Clustree69, selecting a value that induced minimal cluster instability. Datasets were subsetted to include only macrophages, on the basis of expression of key macrophage markers (Adgre1, Csf1r, H2-Ab1, Cd68, Lyz2, Itgam and Mertk). Macrophage-only datasets were then split along conditions and processed anew as described above. DEGs between clusters were identified as those expressed in at least 25% of cells with a log fold change greater than +1 and an adjusted P value of less than 0.01, using the FindMarkers function in Seurat v.4 with all other parameters set to default. Ribosomal protein genes were excluded from the results. Cluster-specific genes were explored for pathway enrichment using StringDB70. DEGs (adjusted P < 0.05, log2 fold change > 0.5) across clusters were subjected to gene ontology pathway enrichment analysis using DAVID71 (v.2016 and v.2021). Gene set scores were calculated using UCell with default parameters72.
Bulk RNA-seq procedure and analysis
RNA-seq data were obtained by sorting 5,000–100,000 cells from each population directly into RLT buffer (Qiagen) containing 1% 2-ME and submitting them to the Emory Integrated Genomics Core at Emory University. Total RNA was isolated using a Quick-RNA Microprep Kit (Zymo Research), and cDNA was generated using a SMART-Seq v.4 Ultra Low Input RNA Kit for Sequencing (Takara Bio) and used to generate sequencing libraries with a Nextera XT kit (Illumina). Libraries were pooled at equimolar ratios and sequenced on a NovaSeq 6000 at approximately 100M reads per sample using 100PE reads. Sequenced libraries were processed for analysis with deepTools73 v.2.0, using STAR65 v.2.7.10 for trimming and mapping and feature Counts74 v.2.0.3 to quantify mapped reads. Raw mapped reads were processed in R (Lucent Technologies)66, using DESeq2 (ref. 75) v.1.36 to generate normalized read counts for visualization as heatmaps with Morpheus (Broad Institute) and to determine DEGs with fold change greater than 1.4 and adjusted P value less than 0.01. Gene ontology analysis was performed with DAVID71 (v.2016 and v.2021).
Ribo-seq pulldown, sequencing and analysis
Freshly isolated cells from total peritoneal lavage were processed as described in ref. 76 with the following modifications. One millilitre of homogenization buffer was used per mouse, and dithiothreitol was removed from the washing buffer to avoid uncoupling of conjugated anti-HA antibodies from magnetic beads. Total lysate (100 μl) was used as an input. The remaining lysate was used for immunoprecipitation of polysomes. Twenty-five microlitres of anti-HA.11 (Pierce) were used per lysate with rotation at 4 °C for 4 h. Beads were then washed three times with 500 μl of wash buffer. RNA was extracted from magnetic beads with TRIzol reagent (Invitrogen), and total RNA was isolated with a Direct-zol RNA Microprep Kit (Zymo Research) according to the manufacturer’s instructions. Total RNA was submitted to Admera Health for RNA-seq. Isolated RNA sample quality was assessed with an RNA TapeStation (Agilent Technologies) and quantified by AccuBlue Broad Range RNA Quantitation assay (Biotium). Paramagnetic beads coupled with oligo d(T)25 were combined with total RNA to isolate poly(A)+ transcripts in a process based on the NEBNext Poly(A) mRNA Magnetic Isolation Module manual (New England Biolabs). Before first strand synthesis, samples were randomly primed (5′ d(N6), 3′ [N = A,C,G,T]) and fragmented on the basis of the manufacturer’s recommendations. The first strand was synthesized with Protoscript II Reverse Transcriptase with a longer extension period, approximately 40 min at 42 °C. All remaining steps for library construction were performed according to the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs). Final library quantity was assessed with Qubit 2.0 (Thermo Fisher), and quality was assessed with TapeStation D1000 ScreenTape (Agilent Technologies). The final library size was about 430 bp with an insert size of about 300 bp. Illumina 8-nt dual-indices were used. Equimolar pooling of libraries was performed on the basis of quality control values, and libraries were sequenced on an Illumina NovaSeq X Plus platform with a read length configuration of 150 paired ends for 40 million paired-end reads per sample (20 million in each direction). For analysis, sequenced libraries were processed as described above for bulk RNA-seq analysis, with the differential expression tests and filters specified in the Results section.
Quantification and statistical analysis
Prism 7 software (GraphPad) was used for statistical analysis, and results are presented as the mean ± s.d., unless otherwise indicated. Comparisons for two groups were performed using unpaired two-tailed Student’s t-tests, and comparisons of more than two groups used one-way analysis of variance with Bonferroni’s multiple comparison tests. Exact P values and details of statistical testing can be found in the figure legends and in the source data file. Unless otherwise specified, n represents the number of individual biological replicates and is represented in graphs as one dot per sample. Flow cytometry plots are representative of at least three replicates, immunoblots of at least two independent experiments, and confocal and immunohistochemistry images of at least three independent biological replicates. No statistical method was used to predetermine sample size, but a minimum of three samples were used per experimental group and condition. Experiments were not randomized.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.