Fragment design
When starting from DNA oligos, the number of junctions needed to assemble a construct depends on the length of the construct and the length of the starting oligos being used. A list of all of the component oligos that compose the Sidewinder fragments used in these experiments is provided in Supplementary Table 1. For an oligo of length L, the maximum bases of coding information for a fragment composed from these oligos (Lc) is L − 2Lb, where Lb is the length of the barcode. Toeholds are then chosen starting maximally from Lc bases away from the previous junction. Hand-designed assemblies standardly use the maximal length fragments and use toeholds from position Lc –10 to Lc but can be shifted to avoid unintended toehold secondary structure. NUPACK-designed assemblies choose a 10-base toehold within the range of position Lc −25 to Lc with an ensemble defect of <0.1 from a secondary-structure-free toehold.
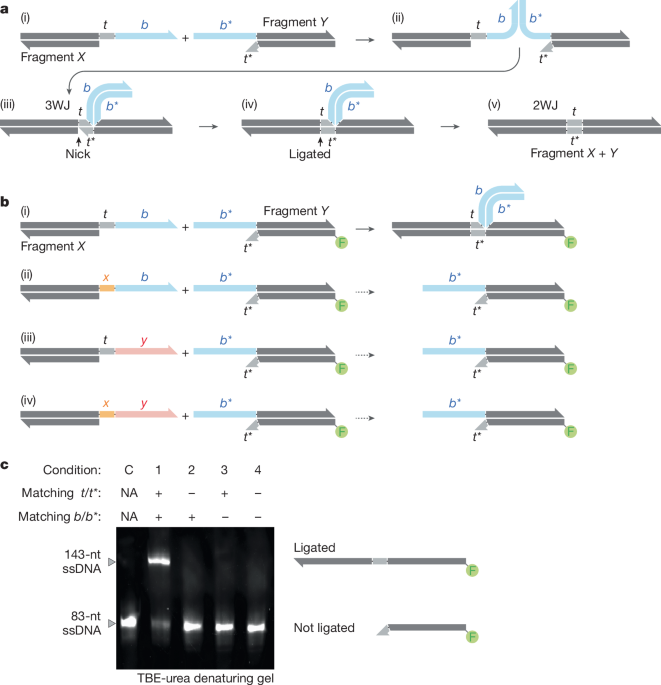
We tested a range of toehold lengths and designs, varying the ligation site from −10 bases to +10 bases on either side of the Sidewinder helix. We found effective ligation occurring equal to or further than ±6 bases from the Sidewinder helix (Extended Data Fig. 2a,b). This led us to standardize the toehold length to 10 bases for the experiments described in this paper as to ensure sufficient distance of the nick from the Sidewinder helix to accommodate ligase docking and effective ligation.
Barcodes were designed to be compatible with their respective toehold after the location of the junction is chosen. Barcode sequences were chosen or generated on the basis of the predicted secondary structure and crosstalk between other toehold–barcode sequences at the assembly’s ligation temperature. The h-fibroin and parallel assembly barcodes were designed using a guess–check method, choosing from a set list of pregenerated orthogonal barcodes15. Starting with the first toehold, a barcode sequence is arbitrarily chosen (‘guess’) and appended to the 3′ end of the toehold and checked for secondary structure at 50 °C with complex size 2 using the NUPACK web browser16,17 (‘check’). The subsequent barcodes are then chosen from the pregenerated list, checked individually in the same manner, then checked for cross reactivity against all previously chosen toehold-barcode sense and antisense sequences at 50 °C and complex size 2. All barcodes in this study use natural bases but we anticipate that the specificity and diversity of Sidewinder barcodes can be expanded to include unnatural bases and other DNA nanotechnology interactions.
The 5-to-40-piece Lux assemblies, the APOE assembly and the library assembly had bespoke barcode sequences generated for the specific assembly using NUPACK Python package16. Target strand (NUPACK variable) secondary structure was defined to be fully unpaired for each single stranded barcode/toehold pair combination. Complexes (NUPACK variable) were defined to take on the desired 3WJ structure for barcode/toeholds. Step tubes (NUPACK variable) are defined such that, in step 0, individual barcode/toehold sequences take on the desired unpaired secondary structure before assembly; and, in step 1, barcode/toehold sequences pair with the intended assembly partner during assembly at 50 °C with an ensemble defect of <0.1. After barcode generation, all secondary structures and the cross-reactivity of chosen barcodes were checked using the NUPACK web browser16,17.
For the length of the Sidewinder barcodes, we tested barcode lengths from 15 to 21 bases, both with and without a T–T or U–U mismatch at the base of the 3WJ for added stability46 and neither seem to have bearing on ligation efficiency (Supplementary Table 1). We also tested a variety of commercially available ligases and found high variability in ligation efficiency at −10 bases from the Sidewinder helix across the ligases tested (Extended Data Fig. 2c). Of the various ligases tested, Taq ligase and HiFi Taq ligase were picked as the preferred ligases due to their efficiency of ligation at the 3WJ and stability at extremely high temperatures.
Oligo purchasing
All assembly oligos were purchased from Millipore-Sigma with standard DNA synthesis for DNA oligos in tubes, which has a max oligo length of 120 bases. The only exception was fragment 4 of the identical toehold assembly, which was ordered as a long oligo to enable four identical toeholds (Supplementary Table 1). Barcode oligos were ordered with standard desalt purification and coding oligos were ordered PAGE purified, but this has since been seen to be superfluous (Extended Data Fig. 3f). Both barcode and coding oligos for the fluorescent protein library were ordered with cartridge purification. Both barcode and coding oligos for the Sidewinder characterization in Fig. 1 were ordered PAGE purified. PCR amplification primers were ordered from Integrated DNA Technologies with standard desalt purity. All oligos are listed in Supplementary Table 1. All Sidewinder component oligos were shipped dry.
Heteroduplex annealing
Oligos are suspended by hand in 1× TE buffer at pH 8.0 (Corning, Thermo Fisher Scientific) to a final concentration of 100 μM on the basis of the manufacturer’s reported weight. To ensure adequate resuspension of the dried oligos, if the volume required to for a final concentration of 100 μM was less than 50 μl of TE buffer according to the manufacturer’s reported weight, the oligos would be resuspended in a volume of 50 μl of buffer, resulting in a lower final concertation. The concentration of all oligos was additionally measured using the Qubit ssDNA Assay Kit (Invitrogen, Thermo Fisher Scientific) and final concentration calculations were based on these measurements.
Sidewinder fragments are generated from resuspended stock oligos by annealing coding oligo to the barcode oligo to form a heteroduplex. To prepare the Sidewinder fragment, the volume of coding oligo required for 2 µM in a 50 µl reaction is first phosphorylated alone in a 25 μl reaction using 1 μl of T4PNK (New England Biolabs) in 1× T4 ligase buffer at 37 °C for 1 h, followed by an enzyme deactivation at 80 °C for 10 min. The corresponding volume of stock barcode oligo needed for 1 μM in 50 μl is then added to the phosphorylated coding oligo and the final volume is topped off to 50 μl using 1× T4 ligase buffer.
Heteroduplexes are then annealed together in a PCR tube consisting of an initial denaturation of 98 °C for 10 min, followed by a gradual decrease in temperature down to 25 °C at −1 °C min−1. Once fragments are annealed, they are kept at 4 °C until use and have been stably used months after initial heteroduplex formation.
Heteroduplex gel extraction
PAGE gel extraction of annealed heteroduplexes is performed using 8% TBE gel (Invitrogen, Thermo Fisher Scientific) and run at 200 V for 35 min. Gel extraction was performed according to a published DNA nanotechnology protocol47.
Sidewinder assembly conditions
Processed Sidewinder fragments are combined into a single reaction mix at equimolar concentrations at around 1 nM to conduct the Sidewinder assembly. We used two avenues for the assembly conditions for the Sidewinder assembly. The assemblies were conducted in 70 μl reactions in 1× HiFi Taq buffer (New England Biolabs). For all assemblies except for the h-fibroin assembly, we used a cycling protocol of 85 °C for 5 min, followed by the addition of 2.8 μl of HiFi Taq ligase (New England Biolabs), then the reaction then cycles between 85 °C for 1 min and 50 °C for 2 min for 100 cycles. These cycles are then followed by 50 °C for 1 h. The second assembly protocol that was used for the h-fibroin assembly was as follows: 13 nM fragments in a 70 μl reaction in 1× HiFi Taq buffer (New England Biolabs) at 85 °C for 5 min, followed by cooling at a rate of −0.1 °C per 6 s down to 50 °C, followed by addition of 2.8 μl of HiFi Taq ligase and then incubation overnight at 50 °C. The Sidewinder characterization assemblies in Fig. 1 were also conducted using the second assembly protocol, with a ramp down to a ligation temperature of 72 °C.
The reactions that characterized choice of ligase and toehold length (Extended Data Fig. 2) used the non-cycling assembly protocol at a ligation temperature according to manufacturer’s recommendation in the corresponding ligase buffer.
Conventional assembly comparison conditions
Fragments for the 4 bp 2WJ, 10 bp 2WJ and Gibson assemblies were generated using oligos. The 4 bp 2WJ and 10 bp 2WJ used the same coding oligo sequence as the corresponding fragment in the Sidewinder assembly. A new complementary oligo was ordered to generate the desired overhangs: a 10 bp 2WJ complement oligo was designed by removing the barcode sequences from the barcode oligo. 4 bp 2WJ was designed to use the terminal 4 bases of the Sidewinder toehold. Gibson oligos were designed to compose an analogous segment with 20 bp of homology to the partner fragment on either end. PCA does not conduct assemblies using fragments but, instead, uses individual oligos that were designed with 20 bases of overlap to the partnered oligo (Extended Data Fig. 3a).
The 4 bp 2WJ, 10 bp 2WJ and Gibson oligos were processed to mirror the Sidewinder fragment processing. Oligos were mixed at an equal 1 µM ratio, both oligos were phosphorylated with T4 PNK (New England Biolabs) in a 50 µl reaction in 1× T4 Ligase buffer (New England Biolabs) and then annealed. Gibson oligos are not phosphorylated. The fragments were PAGE-extracted and the concentrations were measured using the Qubit 1× dsDNA High Sensitivity Assay Kit (Invitrogen, Thermo Fisher Scientific).
The fragments were assembled at the same concentration of the corresponding Sidewinder assembly. The 4 bp 2WJ and 10 bp 2WJ were assembled at 16 °C overnight in 1× T4 ligase buffer with 1 µl T4 Ligase according to the manufacturer’s recommendations for ligating sticky ends (New England Biolabs). Gibson assembly is conducted at 50 °C for 1 h using NEBuilder HiFi DNA assembly Master Mix (NEB). PCA was preformed using PrimeSTAR GXL Polymerase (Takara Bio) under a published protocol that was demonstrated to be optimized for multifragment assemblies48.
PCR amplification and purification
Either PrimeSTAR GXL Polymerase or repliQa HiFi ToughMix (Quantabio) was used for amplification of 3WJ assemblies. Only 1 μl of unpurified 3WJ assembly from the previous ligation step is sufficient template in a 50 μl PCR reaction. PCR reaction conditions were established according to manufacturer recommendations and predicted Tm of primers.
Post PCR amplification, purification of the PCR reaction was done using a QIAquick PCR Purification Kit (Qiagen). Multiple 50 μl PCR reactions can be passed simultaneously through the same purification column to increase the final concentration of the purified 2WJ assembly. Alternatively, gel extraction of the target band can be done. Gel extraction results in an even more highly pure product for downstream sequencing or cloning, as seen with the high-GC assembly (Source data). Gel extraction carried out before sequencing for all assemblies, except for the parallel assembly. The Monarch DNA Gel Extraction Kit (New England Biolabs) was used according to the manufacturer’s protocol.
DNA gel imaging
The Sidewinder characterization gel in Fig. 1 is a 6% TBE-urea denature gel (Novex, Thermo Fisher Scientific) run at a constant 180 V for 30 min in 1× TBE buffer. The samples were mixed with an equal volume of 2× stain free TBE-urea loading buffer and heat shocked at 90 °C for 15 min before loading gel.
All other gel images are 1–2% agarose gels stained with Sybr Safe (Invitrogen, Thermo Fisher Scientific) run at 135 V for 25 min in 0.5× TBE buffer. The 1 kb+ ladder (New England Biolabs) was used in Figs. 2, 3b, 4 and 5. The low molecular mass ladder (New England Biolabs) was used in Fig. 3e. All main figure agarose gels show 50 ng DNA loaded into each lane as measured using the Qubit 1x dsDNA High Sensitivity Assay Kit (Invitrogen, Thermo Fisher Scientific). 4 bp 2WJ, 10 bp 2WJ and Gibson lanes depict 1 µl loaded, as amplification was insufficient to produce 50 ng after purification in some cases.
Cloning and transformation
Sidewinder constructs to be expressed in bacteria were amplified using dU containing primers and repliQa polymerase (QuantaBio). The corresponding vectors were amplified using dU containing primers. The purified products were treated with 1 μl USER (New England Biolabs) in 1× CutSmart buffer and subsequently repurified with PCR Kleen Purification Spin Column (Bio-Rad) and assembled according to published protocols49 in 1× T4 ligase buffer and 2.5 µl T4 ligase. In total, 2 μl of assembled product was electroporated into electrocompetent DH10b cells, recovered in 2 ml Luria–Bertani (LB) medium for 1 h, and plated onto LB-agar plates with the corresponding antibiotics.
All final constructs can be found in FASTA format in Supplementary Table 2.
Sequencing analysis
The final assemblies were processed as described and the purified samples were used for sequencing. Oxford Nanopore Sequencing was used to obtain long, full-molecule reads required to determine the percentage of complete 3WJ assemblies for Figs. 2–4. PacBio sequencing was used to get high-confidence per-base whole-molecule sequencing for the Sidewinder Library in Fig. 5.
Assemblies validated through Nanopore sequencing used Plasmidsaurus Premium PCR Sequencing services. Assemblies validated through PacBio used Azenta sequencing services. To validate our analysis pipeline, each individual raw read for the Sidewinder 40-piece assembly was viewed and assigned manually, enabling us to know the exact identity of each read without any pre-bias due to filtering. For each of the subsequent assemblies, the verified fragment level analysis pipeline is used to generate the pie charts. In this pipeline, read sequences were aligned to fragment references using BLASTn, in which every read was aligned to every fragment reference. The read is assigned as unusable if no hits to any fragment are returned. A correct assembly was assigned when all fragments were in the correct order of the gene sequence. Correct assemblies with all fragments were deemed to be complete, whereas those with not all fragments were deemed to be partial. For Nanopore, all of the remaining reads were checked manually to determine the nature of the assembly. Raw counts can be viewed in the Source data.
The fragment-level analysis has reads sorted into four main categories. A Sidewinder product is a construct that results from the ligation of the toeholds at the 3WJ, characterized by a seamless sequence transition between the 5′ end of one fragment and the 3′ end of another fragment. A correct assembly is a seamless transition between all partnered fragments in the correct order, whereas an incorrect assembly is the seamless transition between non-partnered fragments. In addition to Sidewinder products, PCR artifacts and sequencing artifacts would be expected. PCR artifacts result from mispriming during PCR. These are identified by the sequence transition between two non-partnered fragments joined, not at the assembly junction, but instead at the internal portion of one of the fragments, indicating that a primer or unreacted fragment oligo misprimed and was elongated during PCR (Extended Data Fig. 4a,b). Mispriming can be reduced by biasing the PCR template towards a higher proportion of full-length 3WJ assembly, as well as by using alternative methods for removing the 3WJ that do not depend on PCR amplification (Extended Data Fig. 5a,b). Sequencing artifacts are due to systematic failures in base calling or sequencing preparation procedure that would not result from assembly. Lastly, in protocols in which the barcode oligo is also phosphorylated, we see barcode artifacts where, at a low frequency, the ends of the 3WJ become unintentionally ligated to itself and appear in the final sequence.
For the junction analysis checks, using the same datasets, we conducted an analysis specifically on the junction areas in the reads. A junction is defined as 25 base pairs of both the 3′ and 5′ ends of a Sidewinder junction. For the PacBio data, the number was chosen to be 18 base pairs to avoid degenerate bases being included in junctions. For each sequencing run, we generated a list of all possible Sidewinder junctions, including results of both correct and incorrect ligations, and aligned them to the raw fastq files through BLASTn using sensitive parameters (task=blastn_short -word_size 7 -reward 1 -penalty -3 -gapopen 5 -gapextend 2). The resulting junctions were filtered with bitscore thresholds that were chosen to avoid false-positive BLAST hits while maximally retaining possible misligations. Reads containing misligations are collected and examined manually to verify whether they are true misligations or false positives. The junctions for all sequencing runs were generated and analysed as described above and visualized using custom Python scripts. All junction connections are available in the Source data.
Further analysis of the PacBio sequencing data was conducted to achieve base-level resolution for single-nucleotide polymorphism and diversity analysis of the Sidewinder library. The reads were aligned to reference sequences using the Smith–Waterman algorithm from EMBOSS50 with a match score of 5, mismatch penalty of 4, gap open penalty of 10 and a gap extend penalty of 0.5. To characterize gene-level mutation profiles, reads with incorrect lengths (±20 bp from reference sequence), reads with missing fragments or reads not aligned with target sequence reference are removed from this analysis. Only reads with an average phred score of Q39.5 or above were retained for this downstream analysis and only bases with a phred score of Q40 were included for base-level mutation analysis to minimize errors introduced during sequencing.
Library construction
A sequence of 11 N-degenerate bases was placed before the promoter on the first fragment in the assembly to be able to track individual constructs with defined mutation profiles throughout the assembly, sequencing and transformation. This sequence appears only in the coding oligo and does not have a complement in the barcode oligo.
To ensure efficient retention of diversity required for library construction provided by degenerate bases and additional coding oligos, the library construction was done using a modified protocol prior to assembly. Both barcode oligos and coding oligos were ordered using cartridge purification but otherwise oligo processing followed the same protocol as for other assemblies. For fragments that require multiple coding oligos to cover all mutation profiles, oligos are processed and fragments are annealed in their own tube as if they were distinct fragments. After heteroduplex formation, fragments are not PAGE extracted. The final fragment concentrations are assumed to be the same across each of the fragments at 1 μM heteroduplex. Equimolar fragment concentrations are used in the final Sidewinder assembly. To achieve this, analogous fragments (that is, all F4 fragments with each different coding oligo) are mixed immediately before assembly into a single tube, vortexed, spun down and then this pool is treated as an individual fragment to be added to the assembly mix. Assembly and amplification are then carried out as described.
Pre-clonal sequencing was performed using purified amplicon of the final assembled product. Post-clonal sequencing was conducted by cloning the Sidewinder library assembly as described in the ‘Cloning and transformation’ section. The post-recovery culture was then grown overnight in 500 ml LB with 20 µg ml−1 chloramphenicol. The overnight culture was then miniprepped in batches 10 ml and each elution pooled and sent for sequencing.
Library data presentation
Empirical codon-level mutation profile ratios were determined by calculating the ratio between bases associated with the specified codon choices. The average absolute deviation was calculated by taking the absolute value of the difference between the empirical proportion a codon appears from the theoretical proportion and averaging this difference for all 37 codon options in the library Fig. 5g.
Library coverage was determined by considering the identity of mutated positions for all 17 positions in the gene and assigning this combination to one of the 442,368 variants. To calculate the coverage of every possible combination of N mutants depicted in Fig. 5k, we first determined the number of ways in which we could combine any number of mutation positions. For example, there are 643 ways to combine any 2 mutation positions in this library, 6,971 ways for 3 mutation positions and so on. We then calculated how many of these possible combinations were seen for every N 1–17, where 17 is the coverage for the entire gene library with 442,368 possible variations.
Combinatorial fluorescence library screening
The combinatorial library generated using Sidewinder to introduce mutations into functional protein assemblies was screened by encapsulating transformed clones into hydrogel microparticles using a droplet generator. These encapsulated clones were subsequently analysed using the SONY SH800S FACS system, equipped with four excitation lasers (405 nm, 488 nm, 561 nm and 638 nm) and six detectors capable of detecting emissions ranging from 400 nm to 780 nm. Sorting conditions were optimized by adjusting the gain settings, with the forward scatter set to 1% and the back scatter set to 25%. Detector gain was set to 25% for all channels, except for the FL1 and FL4 channels, which were adjusted to 30%. The sort delay was calibrated to 14 before initiating sorting. Colonies encapsulated in hydrogels were suspended in phosphate-buffered saline pH 7.4 (Gibco, Thermo Fisher Scientific) and analysed at a sample pressure of 4, with an event speed ranging from 200 to 300 events per second. Populations were gated on the basis of their spectral properties and sorted into individual wells of 96-well plates for subsequent expansion and spectral characterization.
We screened over 300 of these sorted clones using a monochromator on the Tecan Infinite 200 Pro using a three-dimensional fluorescence intensity scan from excitation 250 nm to 600 nm and emission from 400 nm to 700 nm. We then chose six clones with potentially unique spectra and cloned them into an p15a pT7 vector and cloned into pTac T7 polymerase strain. The clones were induced overnight in 3 ml at 100 µM IPTG and 10 µg ml−1 tetracycline, centrifuged and resuspended in 50 μl 1× PBS. The final excitation spectra were captured with emission at 560 nm or 620 nm, and excitation from 300 nm to 520 nm or from 400 nm to 580 nm, respectively. The final emission spectra were captured with excitation at either 420 nm or 460 nm, and emission from 460 nm to 680 nm or from 460 nm to 624 nm, respectively.
Overnight cultures grown at 3 ml at 100 µM IPTG and 10 µg ml−1 tetracycline were optical density (OD) normalized to OD 0.2 and 2 μl was spotted onto 100 µM IPTG and 10 µg ml−1 tetracycline LB agar plates. Images were obtained using FITC (470 nm and 525 nm) and TRTC (530 nm and 605 nm) overlayed with Trans on an ECHO Revolve microscope.
Statistics and reproducibility
The experiments depicted were replicated for reproducibility yielding similar results. The ligation in Fig. 1c was repeated three times. The PCA assemblies in Fig. 2c was repeated three times and the Sidewinder assemblies repeated six times. The Sidewinder assembly in Fig. 3b was repeated six times. The Sidewinder assembly in Fig. 3f was repeated four times. The Sidewinder assembly in Fig. 4b was repeated five times. The Sidewinder assembly in Fig. 5c was repeated four times. The ligation in Extended Data Fig. 2a was repeated twice. The ligation in Extended Data Fig. 2b was repeated three times. The Sidewinder assemblies in Extended Data Fig. 2c were repeated twice. The assemblies using conventional technology in Extended Data Fig. 3d were repeated three times. The PCA assemblies in Extended Data Fig. 3e were repeated twice and the Sidewinder assemblies were repeated three times. The Sidewinder assembly in Extended Data Fig. 3f was repeated twice. The digestion of the 3WJ in Extended Data Fig. 5b was repeated twice. The assemblies using conventional technology in Extended Data Fig. 6b and Extended Data Fig. 6c were repeated three times. The transformation of Extended Data Fig. 7a was repeated twice with the 56 non-coloured clones presented screened from one transformation. In Extended Data Fig. 10d multiple 96-well plates were initially screened with 12 samples recloned into an inducible backbone as described. The displayed clones were chosen to represent the diversity of spectra captured. The gel images included are representative of the experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.