Strains and media
Standard methods were used for the culture, sporulation, genetic crossing and manipulation of S. pombe. Strains were generated through transformations or genetic crosses followed by tetrad dissection. Strains used in this study are listed in Supplementary Table 2. Gene deletion strains were obtained from Bioneer haploid deletion library version 4.0, except for tor1 and gad8, which were deleted using pFA6a-natMX6 construct. Most epitope-tagged strains were generated through PCR module-based methods. GFP or GFP-raf1 was cloned in the pDUAL-Pnmt1 or pUC119-Padh1 vector using In-Fusion cloning. Pombe Minimal Glutamate medium supplemented with adenine, uracil, leucine, histidine and lysine (PMG-5S) was used to overexpress protein under the nmt1 promoter. All other experiments were performed in yeast extract-rich medium supplemented with adenine (YEA) at 30 °C unless otherwise specified. For western blotting analysis, the tor2-ts6 mutant was cultured at 26 °C overnight and then transferred to 30 °C for 16 h. For experiments with antifungal agents, cells were treated with 16 mM caffeine, 0.4 mM fluconazole or 0.3 μM clotrimazole in YEA medium.
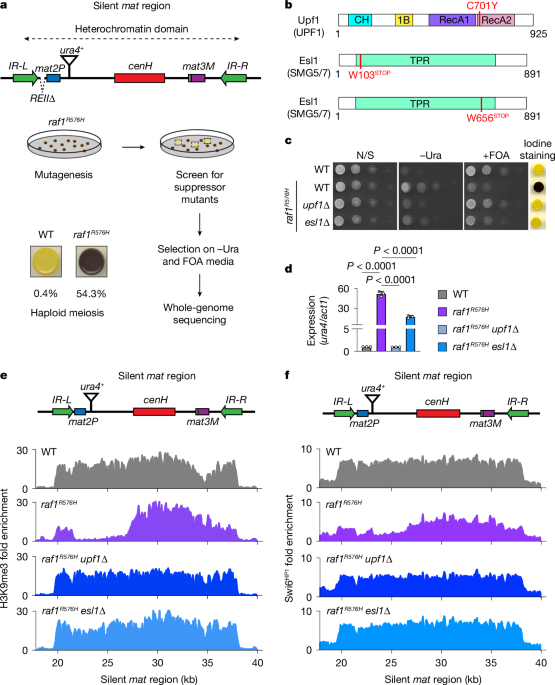
Genetic screen
Genetic screens were conducted using a heterothallic strain (mat1M-smt0) harbouring the raf1R576H mutation and a deletion of the local silencer (REIIΔ) adjacent to the mat2P cassette (Fig. 1a). In addition, the strain contained a ura4+ reporter downstream of mat2P (mat2P::ura4+). Exponentially growing cells were treated with the chemical mutagen methylnitronitrosoguanidine at a concentration of 0.5 mg ml−1 for 60 min at room temperature or irradiated with ultraviolet light (UV) at 100 J m−2 using UV crosslinker. Colonies formed on YEA medium were replica plated onto PMG-5S medium and assayed for haploid meiosis by means of iodine staining. Light colonies were restreaked to single colonies and assessed for growth on −Ura and FOA media. Suppressor mutants were subjected to whole-genome sequencing using the Illumina NextSeq500 platform. Genomic sequencing reads were quality trimmed using fastp61 and aligned to the S. pombe ASM294v2.30 reference sequence62 with the BWA aligner63 using default parameters. Duplicate reads in the resulting BAM files were marked using Picard tools (http://broadinstitute.github.io/picard/) ‘MarkDuplicates’. Mutations were called from the duplicate-marked BAM files using samtools ‘mpileup’ and subsequently processed with bcftools64 to generate a single VCF file65 containing mutations identified in the WT and mutant genomes. Mutation impacts were predicted using SnpEff66. Variations that were present in the mutants but absent in the WT controls and were predicted to have ‘HIGH’ or ‘MODERATE’ impact were flagged for further investigation.
Serial dilution assay for heterochromatic silencing
To assess the silencing of the ura4+ reporter in mat1M-smt0 REIIΔ mat2P::ura4+ cells, tenfold serial dilutions of cultures were spotted on uracil-deficient medium and PMG-5S medium supplemented with FOA. The derepression of mat2P was evaluated by exposing the cells to iodine vapour. Cells undergoing haploid meiosis on PMG-5S medium, which results from mat2P derepression, accumulate a starch-like compound and stain dark brown when exposed to iodine vapour. Images of serial dilution plates were presented with Adobe Photoshop v.22.4.2 and Adobe Illustrator v.2024.
RT–qPCR
Total RNA was extracted from 2 × 108 cells using the MasterPure Yeast RNA Purification Kit (Biosearch Technologies) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 1 μg of DNase-treated RNA using SuperScript III reverse transcriptase with gene-specific primers. The resulting cDNA was subjected to quantitative PCR (qPCR) using iTaq Universal SYBR Green Supermix (Bio-Rad), following the manufacturer’s instructions. The act1+ gene served as the internal control for normalization. Oligonucleotides used for qPCR are listed in Supplementary Table 3.
ChIP–qPCR
For the H3K9me3 ChIP assay, 5 × 108 cells were crosslinked with 3% paraformaldehyde (PFA) for 30 min at room temperature, followed by quenching with 125 mM glycine. For Swi6 or Raf2 or H3K14ub ChIPs, 1 × 109 of cells were incubated for 2 h at 18 °C, fixed in 3% PFA and washed in ice-cold PBS. Cells were treated with 10 mM dimethyl adipimidate at room temperature for 45 min.
Fixed cells were resuspended in ChIP lysis buffer (50 mM HEPES/KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with 1 mM PMSF and protease inhibitors (cOmplete Mini Protease Inhibitor Cocktail, Roche) and lysed by bead-beating (Biospec Mini-Beadbeater-16). For H3K14ub, ChIP lysis buffer was also supplemented with DUB inhibitor 100 μM PR-619 and 100 mM N-ethylmaleimide. The lysates were sonicated with a Diagenode Bioruptor for 14 cycles on medium power setting (30 s ON, 30 s OFF) to shear DNA into 0.4–0.6 kilobase (kb) fragments. Cellular debris was removed by centrifugation and 2% of the supernatant was kept for whole cell extract input control. The remaining lysates were precleared with protein A and protein G agarose beads for 1 h at 4 °C. After bead removal, lysates were incubated with the appropriate antibody (2–10 μg) overnight at 4 °C with slow rotation. Antibodies used include anti-H3K9me3 (Abcam), anti-c-Myc (Santa Cruz), anti-H3K14ub, as described in ref. 37, and custom affinity-purified anti-Swi6. Prewashed protein A and protein G agarose beads were added to the lysates and incubated for 4 h at 4 °C. The beads were sequentially washed, twice with wash buffer I (50 mM HEPES/KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), twice with wash buffer II (50 mM HEPES/KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), twice with wash buffer III (10 mM Tris/HCl pH 8.0, 250 mM LiCl, 0.5% IGEPAL, 0.5% sodium deoxycholate, 1 mM EDTA) and once with TE buffer (50 mM Tris/HCl pH 8.0, 10 mM EDTA). The chromatin was eluted from the beads twice with 50 μl of elution buffer (50 mM Tris/HCl pH 8.0, 10 mM EDTA, 1% SDS) at 65 °C for 30 min each, with shaking (roughly 1,200 rpm). Eluted chromatin and input control samples were decrosslinked overnight at 65 °C. Samples were treated sequentially with RNase A (10 μg, 2 h, 37 °C) and Proteinase K (20 μg, 2 h, 37 °C), and subsequently purified using the Qiagen PCR purification kit. qPCR was conducted with iTaq Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Fold enrichment was calculated using the ΔΔCt method, with leu1+ serving as the reference. Oligonucleotide sequences for qPCR are provided in Supplementary Table 3.
ChIP–seq and data processing
Genome-wide analysis of ChIP DNA was conducted as previously described in ref. 9. One ng of DNA was used for library preparation using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s instructions. Libraries were purified with AMPure XP magnetic beads (Beckman Coulter), and their quality was analysed on the TapeStation System 4150 (Agilent) before sequencing using the Illumina MiSeq sequencing platform. Sequenced reads were quality trimmed with fastp61 and aligned with the BWA aligner63 to the S. pombe ASM294v2.30 reference sequence62 to which we added a separate contig containing the sequence of the full 40-kb mat region. Both the Pombase provided mat region on chromosome 2 (chr. II:2109748–2138781) and the mat1M locus on the 40-kb mat contig (MAT 4,489–5,615) were masked from alignment. ChIP–seq alignments for strains lacking the REII element in the mat region were made to the standard genome as described but with the sequence of the REII element removed. Bedgraphs of ChIP enrichment over input were produced using the MACS2 (ref. 67) ‘callpeaks’ function to make broad calls with options ‘-nomodel–extsize 147’, followed by the MACS2 ‘bdgcmp’ function to compute fold enrichment over the input background. As the sequence of the constitutively euchromatic mat1 locus is identical to that of its counterpart within silent mat3, the euchromatic copy of mat1 was masked when aligning reads to prevent read misalignment. The Integrative Genomics Viewer68 was used to plot ChIP–seq data.
RNA-seq analysis
Transcriptome analysis was performed as previously described in ref. 30. Briefly, total RNA was isolated using a MasterPure Yeast RNA Purification Kit (Lucigen) and then ribosomal RNA (rRNA) was removed using a Ribo-Zero magnetic gold rRNA removal kit (yeast; Illumina). The library was constructed using a ScriptSeq v.2 RNA-seq library preparation kit (Illumina) or NEBNext Ultra II directional RNA library prep kit for Illumina (NEB). The final library was analysed using an Agilent 2100 BioAnalyzer and sequenced on the Illumina NextSeq500 platform. Single ended short reads from RNA-seq experiments were quality trimmed using fastp61 and aligned using the STAR aligner69. Variable interval bedgraphs, normalized to counts per million mapped reads and including both uniquely and multi-mapping reads, were generated by STAR and were further processed to produce 10-base pair fixed interval versions for purposes of figure construction. Read counts for transcripts were computed from BAMs using Rsubread70 and differential expression with respect to WT controls was computed using DEGseq71. The Volcano plot was constructed from log2 fold-changes and P values produced by DEGseq using the ‘EnhancedVolcano’ R library (https://github.com/kevinblighe/EnhancedVolcano).
Northern blotting
Northern blot analysis of centromeric small interfering RNAs (siRNAs) was performed as described previously in ref. 72. Briefly, small RNAs (less than 200 nt) were purified from mid-log phase cells with mirVana microRNA isolation kit (Thermo Fisher Scientific). Then 20 μg of small RNAs were resolved on a 15% denaturing acrylamide gel and transferred to Hybond-N+ (Thermo Fisher Scientific) membrane in 0.5× TBE for 1 h at 100 V. After UV crosslinking, the membrane was hybridized with α-P32-UTP (PerkinElmer) labelled RNA probes (roughly 50 nucleotides) corresponding to the dg sequence in ULTRAhyb-Oligo hybridization buffer (Thermo Fisher Scientific). The membrane was exposed and scanned using Typhoon FLA 9500 phosphor imager (GE Healthcare).
For northern blot analysis of raf1 transcript, exponentially growing cells were resuspended in LETS buffer (100 mM LiCl, 10 mM EDTA, 10 mM Tris/HCl pH 7.5, 0.2% SDS), combined with equal volume of LETS-saturated phenol–chloroform and lysed by bead-beating. Total RNA was purified through repeated extraction with LETS-saturated phenol–chloroform and collected by precipitation after the addition of LiCl to a final concentration of 0.5 M and 2.5 volumes of 100% ethanol. The RNA was resuspended in NorthernMax formaldehyde loading dye (Invitrogen), resolved on a 1% agarose gel and transferred onto a BrightStar-Plus (Invitrogen) membrane by following the NorthernMax kit (Invitrogen) protocol. Blotted RNA was detected using α-P32-UTP (PerkinElmer) labelled RNA probe. RNA probe was prepared by in vitro transcription with the Maxiscript T7 kit (Invitrogen). Probe was hybridized to blotted RNAs in ULTRAhyb hybridization buffer (Invitrogen) and the blots were visualized using the Typhoon FLA 9500 phosphor imager (GE Healthcare).
RIP–qPCR
For the Myc-tagged Upf1 RIP assay, 1 × 109 cells were incubated for 2 h at 18 °C, fixed in 3% PFA and washed in ice-cold PBS. Cells were treated with 10 mM Dimethyl adipimidate at room temperature for 45 min. Fixed cells were resuspended in ChIP lysis buffer (50 mM HEPES/KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with 1 mM PMSF, complete EDTA-free proteinase inhibitor cocktail (Roche) and 40 units of RNase inhibitor (Thermo Fisher Scientific, AM2694) and lysed by bead-beating (Biospec Mini-Beadbeater-16). The lysates were sonicated with a Diagenode Bioruptor for 14 cycles on medium power setting (30 s ON, 30 s OFF). Cellular debris was removed by centrifugation, and 5% of the supernatant was reserved for whole cell extract input control. The remaining lysates were precleared with 0.9 mg of prewashed protein G Dynabeads (Invitrogen, 10004D) at 4 °C for 1 h. After bead removal, lysates were incubated with 5 μg of anti-c-Myc antibody (Santa Cruz, 9E10) overnight at 4 °C with slow rotation. Antibody–protein complexes were captured using 1.2 mg of protein G Dynabeads for 2 h at 4 °C. The beads were sequentially washed in 900 μl of each buffer, once with wash buffer I (50 mM HEPES/KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), once with wash buffer II (50 mM HEPES/KOH pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), once with wash buffer III (10 mM Tris/HCl pH 8.0, 250 mM LiCl, 0.5% IGEPAL, 0.5% sodium deoxycholate, 1 mM EDTA) and once with TE buffer (50 mM Tris/HCl pH 7.0, 10 mM EDTA). Each wash was done for 7 min at slow rotation. Beads were eluted twice in 75 μl of RIP elution buffer (50 mM Tris/HCl pH 8, 10 mM EDTA, 300 mM NaCl, 1% SDS) at 37 °C for 10 min. To 50-μl input samples, 100 μl of RIP elution buffer was added to make a final volume of 150 μl. Then 20 μg of proteinase K (Thermo Fisher Scientific, AM2548) was added to both immunoprecipitation and input samples, and the mixtures were incubated at 37 °C for 1 h followed by de-crosslinking at 65 °C for 1 h. The samples were then extracted with phenol–chloroform, precipitated with ethanol and the pellet was resuspended in 80 μl of DEPC-treated water. The samples were further treated with 20 units of RNase-free DNase I (Thermo Fisher Scientific, AM2222) at 37 °C for 1 h, extracted with phenol–chloroform and ethanol precipitated as above. Immunoprecipitation and input samples were resuspended in 20 µl and 50 µl water, respectively. RNA was used for performing RT–qPCR as mentioned above. Fold enrichment was calculated using the ΔΔCt method, with act1+ serving as the reference. Oligonucleotide sequences for qPCR are provided in Supplementary Table 3.
Western blotting
The 4 × 107 mid-log phase S. pombe cells were lysed in 20% trichloroacetic acid by bead-beating (Biospec Mini-Beadbeater-16). The precipitated proteins were washed with ethanol. For Flag-Clr4, cells were resuspended in 150 μl TBS (50 mM Tris/HCl pH 7.5, 150 mM NaCl), heated for 5 min at 95 °C and lysed by bead-beating. Lysate was resuspended in SDS–PAGE sample buffer, and resolved using polyacrylamide gel electrophoresis. Anti-c-Myc 9E10 (Santa Cruz, sc-40), anti-Flag M2 (Sigma Aldrich, F3165) and mouse IgG HRP-linked (GE Healthcare, NA931) antibodies were used for probing the epitope-tagged proteins. For endogenous proteins, affinity-purified anti-Swi6 (in-house), antibody specific for Gad8 phosphorylated at Ser546 (gift from R. Weisman) and Rabbit IgG HRP-linked (GE Healthcare, NA934) antibodies were used. Primary antibody dilution was 1:1,000 and secondary antibody dilution was 1:2,500.
Glycerol-gradient fractionation
The 1 × 109 S. pombe cells grown to mid-log phase were pelleted and flash frozen in liquid nitrogen. The frozen cells were lysed using a CryoMill (Retsch) with the parameters: three cycles at 30 Hz for 1 min followed by 5 Hz for 1 min, and the resulting cell powder was resuspended in lysis buffer (20 mM Tris/HCl, pH 8.0; 2 mM EDTA; 1% IGEPAL; 2 mM β-mercaptoethanol; 137 mM NaCl and 2× complete proteinase inhibitor). The lysate was centrifuged at 27,000g at 4 °C for 30 min, and protein concentrations of the supernatant were measured using the Bradford assay. A discontinuous glycerol gradient (1.8 ml) was prepared by layering glycerol solutions with decreasing concentrations (200 μl each of 50%, 45%, 40%, 35%, 30%, 25%, 20%, 15% and 10% glycerol) in lysis buffer within a polyallomer ultracentrifuge tube (Beckman Coulter). The gradient was allowed to form at room temperature for 2 h, followed by cooling at 4 °C for an extra 2 h. Subsequently, 120 μl of whole cell extract was loaded atop the gradient. The tubes were inserted into precooled buckets and centrifuged at 35,000 rpm in a Beckman TLS-55 rotor at 4 °C for 19 h. Fractions (150 μl) were collected starting from the top of the gradient, and 10 μl from each fraction was resolved on a 4–12% SDS–PAGE Bis-Tris gel, followed by western blotting to detect specific proteins.
Immunoprecipitation analysis
A 2-l culture of S. pombe was harvested at an optical density at 600 nm (OD600) of 1. Cells were washed with ice-cold distilled water and resuspended in 2 ml of lysis buffer (300 mM HEPES/KOH pH 7.6; 100 mM KCl; 2 mM EDTA; 2 mM PMSF; 1 mM DTT (dithiothreitol); 0.2% IGEPAL; Roche complete mini protease inhibitor cocktail) and flash frozen in liquid nitrogen as nuggets. The frozen cell nuggets were then lysed using a CryoMill (Retsch) with the following parameters: nine cycles at 30 Hz for 1 min followed by 5 Hz for 1 min. The resulting cell powder was resuspended in 10 ml of lysis buffer. The lysate was centrifuged at 4,000 rpm for 10 min, and the supernatant was further clarified by ultracentrifugation at 27,000g for 1 h at 4 °C. Two hundred microlitres of M2-Flag beads (Sigma Aldrich, A2220) were washed three times with wash buffer (150 mM HEPES/KOH pH 7.6, 250 mM KCl, 1 mM EDTA, 1 mM PMSF, 0.5 mM DTT, 0.1% IGEPAL) and subsequently added to the clarified lysate. After 2 h of incubation at 4 °C, beads were washed twice with wash buffer, followed by two washes with AC buffer (20 mM HEPES/KOH, pH 7.6, 200 mM KCl, 1 mM EDTA, 2 mM MgCl2, 0.5 mM DTT, 0.1% IGEPAL). Beads were resuspended in 200 μl of AC buffer containing 500 μg ml−1 Flag peptide for elution of bound protein. After overnight incubation at 4 °C, proteins in the eluate were precipitated by adding 10% trichloroacetic acid on ice for 10 min. The protein pellet was washed with acetone, air-dried, resuspended in 200 μl of SDS–PAGE sample buffer, boiled at 95 °C for 5 min and analysed by western blotting.
Ectopic heterochromatin silencing assay
The reporter strains contain 6xtetO-ade6+ inserted at the ura4+ locus and Pnmt81-tetR-2xflag-clr4+ integrated at the leu1+ locus. Strains were grown in Edinburgh Minimal Medium (EMM) liquid media for 48 h to induce TetR-Clr4 expression. Expression and heterochromatization of the ade6+ reporter were assayed by serial dilution and ChIP–qPCR analyses, respectively. Tenfold serial dilutions were spotted on low-adenine (7.5 mg l−1) EMM medium lacking thiamine (tetR-clr4ON) or supplemented with 5 mg l−1 thiamine (tetR-clr4OFF), and ade6+ expression status was determined by observing colony coloration. White colonies indicate ade6+ expression, whereas ade6+-repressed cells form red colonies. H3K9me was analysed in cells grown for 48 h in EMM and collected at OD600 of 0.5 (establishment, tetR-clr4ON cells). For heritability of H3K9me, cells were further cultivated in liquid EMM supplemented with tetracycline (2.5 mg l−1) and thiamine (5 mg l−1) for 10 generations and fixed with 3% PFA at OD600 of 0.5. Fixed cells were processed for ChIP as described earlier. Oligonucleotides used for qPCR are listed in Supplementary Table 3.
Live-cell imaging
Cells were grown overnight at 26 °C in PMG-5S medium until they reached logarithmic phase. For Halo tagged proteins, cells were stained with 25 nM Janelia Fluor 646 HaloTag ligand (Promega) for 1 h. For mat2P::GFP imaging, YEA medium was used. Cells were mounted on a 2% agarose pad formed on a glass slide. Images were acquired on a DeltaVision Elite microscope (Leica) with a ×100, 1.35 numerical aperture oil lens (Olympus). Optical z sections were acquired (0.2-μm step size, 20 sections) for each field. Images were deconvolved and all z-stacks were projected into a single-plane as maximum-intensity projections. Fiji (ImageJ2 v.1.53, National Institutes of Health (NIH)) was used for processing the images.
Image acquisition for SMT
Cells were grown in PMG-5S medium to mid-log phase and stained with 100 pM of Janelia Fluor JFX650 HaloTag ligand (JFX650 Promega) for 1 h at 30 °C. Cells were then briefly washed twice, resuspended in fresh media, mounted on a chambered coverglass (Lab Tek II) and covered by a 1% agarose patch. Imaging was performed on custom-built HiLO microscope73. This custom-built microscope from the CCR, LRBGE Optical Microscopy Core facility is controlled by μManager v.2.0 software (Open Imaging Inc.), equipped with an Okolab state top incubator for temperature control (30 °C) with an objective heater, a ×100, 1.49 numerical aperture objective (Olympus Scientific Solutions), 647-nm and 488-nm lasers (Coherent OBIS), and EM-CCD cameras (Evolve 512 Delta, Photometrics). Images for GFP and JFX650 (for HaloTag) were acquired simultaneously with an exposure time of 100 ms, and a time-lapse interval of 200 ms for a total of 2 min, with laser powers of 3 mW for the 647-nm laser and 100 mW for the 488-nm laser.
SMT analysis
The custom-made software TrackRecord74 in MATLAB (v.R2024a, The MathWorks Inc.) was used for analysis (https://sourceforge.net/projects/single-molecule-tracking/). Briefly, to analyse each time series, data were filtered using top-hat, Wiener and Gaussian filters. A region of interest was defined to encompass the nuclei based on the nuclear GFP staining, then nuclear particles were detected, fitted to two-dimensional Gaussian function for subpixel localization and finally tracked using a nearest neighbour algorithm75. The tracking parameters were as follows: window size for particle detection 3 pixels, maximum frame-to frame displacement of 6 pixels, shortest track 2 frames and gaps to close 2. Diffusion coefficients of individual tracks were estimated by calculating the mean-squared displacement over time and fitting the first four points in the curve to a line, the slope of which gives the average diffusion coefficient. Only those fits whose confidence intervals included only positive numbers were retained for the analysis.
Track segments were classified into distinct diffusive states using pEM (v.2)76. Assumptions about the type of diffusion shown by the tracked particles are not required with pEM and the number of diffusive states can be deduced from the analysis. pEM analysis requires all analysed tracks to be of the same length, and it is preferable to use shorter tracks that do not include transitions between states. Therefore, tracks were split into seven frame segments and the pEM classification analysis was performed on the set of all these track segments. The minimum number of states for the system to converge to was set at two and the maximum at seven. If the optimal number of states that the analysis converged to was seven, the algorithm was rerun with a higher number of maximum states. The number of reinitializations was set to 50 with 200 perturbation trials. The maximum number of iterations was 1,000 with a convergence criterion for the change of log likelihood of 10−7. The number of features for the covariance matrix was set to three for tracks of length seven. A motion blur coefficient was calculated as (1/6)(Δe/Δt), where Δe corresponds to the exposure time and Δt the acquisition interval.
Survival distributions were calculated from the immobile portions of particle tracks as in ref. 77. Immobile segments of tracks were identified on the basis of their displacements compared with HaloTag-H2B data, which was acquired with identical imaging conditions to the Raf2. Because the motion of H2B includes the thermal motion of chromatin, it can be used to determine the maximum distance a DNA-bound molecule will be allowed to move. Therefore, we used the jump distribution of H2B tracks to define three parameters: Rmin, Rmax and Nmin. Rmin is the maximum distance for which histone molecules can move between two consecutive frames, calculated as the 99th percentile for H2B frame-to-frame displacements. Rmax is the maximum distance for which bound histone molecule may move in Nmin frames, calculated as the 99th percentile of displacements of histone molecules after Nmin frames. Nmin is the minimum frame number of a shortest bound track, and is calculated to obtain a low probability, P = 0.01, that a diffusing molecule is classified as bound using the following equation:
$$P({R}_{\text{min}},\,{N}_{\text{min}})=(1-{{\rm{e}}}^{-\frac{{R}_{\text{min}}^{2}}{4D\Delta t}})\genfrac{}{}{0ex}{}{{N}_{\text{min}}}{\,}$$
The photobleaching rate was estimated as the slowest exponential decay obtained from fitting the H2B survival distribution to a triple exponential decay. The survival distribution of the Raf2 was corrected for photobleaching as follows (\(S(t)={\rm{SE}}(t)/\gamma (t)\), where S(t) corresponds to the survival distribution after photobleaching correction, SE(t) the empirical survival distribution and γ(t) is the normalized photobleaching decay).
In vitro affinity pulldown
Recombinant His6-Clr4, glutathione S-transferase (GST) and GST-Raf1 (amino acids (aa) 211–638) were expressed and purified from Escherichia coli (BL21). His6-Clr4 was purified by Ni-bead affinity pulldown followed by high-performance liquid chromatography size fractionation (GenScript). GST and GST-Raf1 (aa 211–638) were purified by affinity pulldown using Glutathione Sepharose 4B resin (Cytiva, 17075601). Purified His6-Clr4 was added to GST or GST-Raf1 (aa 211–638) bound Glutathione Sepharose beads in affinity binding buffer (20 mM Tris/HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.05% NP-40, 2 mM DTT and complete protease inhibitors) at a protein ratio of 4:1. After 60 min of incubation with gentle rotation at 4 °C, beads were pulled down by a short spin at 500g. Beads were washed twice with 500 µl of affinity binding buffer and resuspended in SDS sample buffer. Bound proteins were analysed by SDS–PAGE and western blotting for Clr4 with an affinity-purified rabbit α-Clr4 antibody (dilution 1:1,000) and Alexa-Fluor 647 labelled secondary antibody (Invitrogen, A21246) (dilution 1:2,500). Fluorescence signals were captured by scanning using Typhoon FLA 9500 (GE Healthcare).
Quantification and statistical analysis
Quantification and statistical tests used are described in the figure legends. The n represents the number of independent biological replicates. GraphPad Prism v.10 software was used to plot all the graphs and calculate statistical significance.
Materials availability
Strains and plasmids generated in this study are available from the corresponding author upon reasonable request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.