C. elegans husbandry
All C. elegans strains (Supplementary Table 1) were maintained at 20 °C on nematode growth medium (NGM) plates seeded with E. coli OP50 according to previously described methods51, unless otherwise specified. Strains obtained from the Caenorhabditis Genetics Center are listed in Supplementary Table 1. WT refers to the N2 Bristol strain. All strains are available upon reasonable request. All experiments were performed with age-matched C. elegans hermaphrodites. Sample sizes were not predetermined by statistical methods. Randomization was not applied, as experiments were designed according to genotype or condition. Investigators were not blinded to allocation, but control and experimental samples were processed in parallel under identical conditions. No ethical approval is required to work with C. elegans.
Cell lines
HEK293T cells (CRL-3216; sourced and authenticated by the American Type Culture Collection) and HEK293T-derived reporter and knockout cell lines (Supplementary Table 1) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, 11995-065) supplemented with 10% fetal bovine serum (FBS) (Gibco, 10437-028). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. All cells used in this study were tested negative for mycoplasma contamination, and no commonly misidentified cell lines were used.
RNAi
Feeding RNAi
Gravid adults were treated with alkaline-bleach solution (20% commercial bleach, 0.5 M NaOH) to isolate embryos (egg preparation). The embryos were then transferred to RNAi plates (NGM plates containing 1 mM IPTG and 25 mg ml–1 carbenicillin) seeded with E. coli HT115 expressing either control dsRNA (L4440 empty vector) or dsRNAs targeting specific genes (utp-20 or dpy-6). The utp-20 and dpy-6 RNAi colonies are from the Ahringer library and all plasmid identities were confirmed by Sanger sequencing and whole plasmid sequencing (Supplementary Table 1).
esiRNA treatment
Dried esiRNA oligonucleotides targeting RTCB (Sigma Aldrich, EHU009301-20UG) were resuspended in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA). esiRNA targeting RLUC (Sigma Aldrich, EHURLUC-50UG) was used as a negative control in all RNAi experiments. HEK293T cells were transfected with esiRNAs using Lipofectamine RNAiMAX transfection reagent (Invitrogen, 13778075), following manufacturer’s protocols. Transfections were performed at a final concentration of 10 pmol in a 24-well plate. Knockdown efficiency of target genes was confirmed by immunoblot analysis.
In vitro RNA synthesis
To prepare in vitro RNAs as controls for nanopore long-read sequencing experiments, RNAs were synthesized using a MEGAscript T7 Transcription kit (Invitrogen, AM1334). DNA fragments were cloned into a pDONR221 plasmid (Tc1::rsd-3, rsd-3, Tc1::Ngfp) containing the T7 promoter using Gateway BP Clonase II enzyme mix (Invitrogen, 11789-100) or pUC57 plasmid (Tc1-traΔ::rsd-3, Tc1-traΔ::ITRscr::rsd-3) using AccI (NEB, R0161S) and AvaI (NEB, R0152S), linearized and purified using phenol–chloroform extraction to serve as DNA templates for in vitro transcription. Transcription reactions were incubated overnight at 37 °C. Resulting RNA was purified using RNA Clean & Concentrator-5 (Zymo Research, R1014) and stored at −80 °C.
Long-read nanopore sequencing and SOS splicing analysis
Total RNA was extracted from animals using RNA Clean & Concentrator-5 (Zymo Research, R1014), followed by DNase treatment to deplete gDNA contamination. For cDNA synthesis, 1 μg of in vivo RNA (or 1 ng of in vitro RNA) was reverse-transcribed using Induro reverse transcriptase (NEB, M0681L) with random primer mix (60 μM) (NEB, N0447). In brief, RNA was mixed with random primer mix and dNTPs, denatured at 65 °C for 5 min and then immediately chilled on ice. Induro RT reaction buffer, RNase inhibitor and Induro reverse transcriptase were added to the mixture. The reaction was incubated in a thermocycler with the following conditions: 2 min at 25 °C, followed by 30 min at 60 °C and a final hold at 95 °C for 2 min.
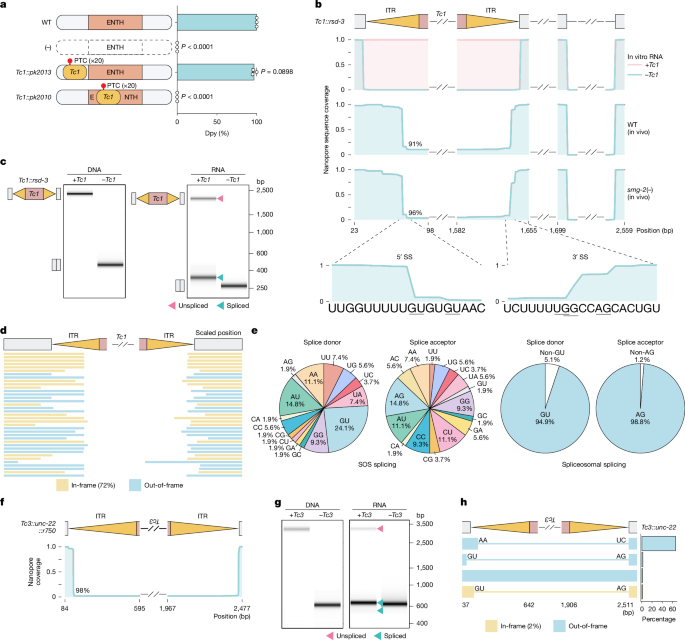
SOS splicing isoforms were amplified using primers flanking transposon insertion sites. In brief, 200 ng cDNA (from in vivo RNA) or 10−5 pg cDNA (from in vitro RNA) was used for PCR amplification. Reactions were performed in a 200 μl reaction volume with 20–23 cycles using Q5 High-Fidelity DNA polymerase (NEB, M0491L). PCR products were then purified using 0.7× to 1× AMPure XP reagent (Beckman Coulter, A63881) depending on the amplicon size and analysed by automated electrophoresis using a 4200 TapeStation system (Agilent, G2991BA). PCR sequencing was performed by Plasmidsaurus using Oxford Nanopore Technologies followed by custom analysis and annotation.
Reads were mapped to the indicated reference genomes (Supplementary Table 2) using Minimap2 (v.2.22, –r1109dirty)52, with specific alignment parameters optimized for SOS splicing events (minimap2 -ax splice -C 0) and for hybrid spliceosome–SOS splicing events and the F19B2.5.2 5′ UTR locus (minimap2 -ax splice). Isoform identification was performed using IsoQuant (v.3.5.0)53 with the following parameters: isoquant.py –data_type assembly –check_canonical –keep_tmp –stranded none –report_canonical all –splice_correction_strategy none –model_construction_strategy sensitive_ont –report_novel_unspliced false. Nanopore sequencing coverage for specified genomic regions was normalized to the highest value, and splicing isoforms and splice sites were identified based on detected splicing events. Spliceosomal splice sites were identified from 10,000 randomly selected C. elegans introns using the annotation file WBcel235.gtf (Ensembl Release 104, WormBase WS276). Results were visualized using R (v.4.4.2).
Automated electrophoresis using TapeStation
Total RNA was extracted from animals using a RNA Clean & Concentrator-5 Kit (Zymo Research, R1014) and reverse-transcribed with Induro reverse transcriptase (NEB, M0681L) using a random primer mix, as described above. For PCR amplification, 50–200 ng of total DNA (to detect transposons in gDNA) or 25 ng of cDNA (to detect transposons in mRNA) was used as templates. Amplification was performed using Q5 High-Fidelity DNA polymerase (NEB, M0491L) for 30 cycles. PCR amplicons were analysed using a 4200 TapeStation system (Agilent, G2991BA) with D1000 ScreenTape (Agilent, 5067–5582) or D5000 ScreenTape (Agilent, 5067–5588) following the manufacturer’s instructions. All primers used in this study are listed in Supplementary Table 1. Uncropped TapeStation results are provided in Supplementary Fig. 1.
SOS splicing reporter microinjection
The Tc1::NmScarlet SOS splicing reporter plasmid (Prpl-28::Tc1::NmScarlet::unc-54 3′ UTR) or variant constructs were microinjected at 50 ng μl–1, along with 2 ng μl–1 of the co-injection marker pCFJ421 plasmid (Pmyo-2::gfp::h2b; Addgene, 34876). Pharyngeal GFP expression (co-injection marker) was used to detect successful microinjection. P0 animals were microinjected into the germline and allowed to lay a brood at 20 °C. F1 animals with pharyngeal GFP expression were scored for mScarlet expression.
EMS screening and whole-genome sequencing
Tc1::rsd-3 (that is, pk2013);Tc1::Ngfp-containing L4 stage animals were washed twice with M9 buffer and then mutagenized with 47 mM EMS by rotating at 20 °C for 4 h. Mutagenized animals were grown on 100 mm NGM plates seeded with E. coli OP50 until most F1 animals had reached the young adult stage. F2 embryos were obtained via egg preparation from gravid adult hermaphrodites and placed on RNAi plates seeded with E. coli HT115 bacteria expressing utp-20 dsRNA. Animals that did not respond to utp-20 RNAi (that is, they developed to young adults) were transferred to NGM plates seeded with E. coli OP50. Lineages established from these animals were tested for GFP expression. All potential mutants resistant to utp-20 RNAi and that did not express GFP were further examined for reporter SOS splicing patterns using RT–PCR as described above.
A mapping strain was established from an animal mutagenized with EMS but exhibited a WT response to utp-20 RNAi (larval arrest) and normal GFP expression. To map mutants, males from the mapping strain were crossed with mutant hermaphrodites, and eggs from F1 adult animals were collected and grown on utp-20 RNAi plates. F2 animals that did not respond to utp-20 RNAi were scored for GFP expression. F2 animal lineages that exhibited SOS splicing defects were pooled (>20 F2 animal lineages per mutant) in nearly equal proportions and processed for gDNA extraction and whole-genome sequencing at the Biopolymers Facility at Harvard Medical School or BGI Genomics. Reads were aligned to the C. elegans genome (WBcel235/ce11) using BWA-MEM (v.0.7.17-r1188), and variants were identified using Samtools (v.1.3.1) and bcftools (v.1.13). Mapping single-nucleotide polymorphisms were genotyped using GATK (v.4.1.9.0), and unique mutations absent in the mapping strains were plotted using R (v.4.4.2).
Microscopy imaging of C. elegans
For live-worm imaging, worms were collected from NGM plates and washed once with M9 buffer. To restrain worm movement, 2% low melting agarose (Invitrogen, 16520050) was mixed with animals and then seeded onto glass slides. After the agarose solidified, imaging was performed using a Nikon Ti2 W1 Yokogawa spinning disk confocal microscope equipped with a Plan Apo λ ×20/0.8 DIC I or a Plan Apo λD ×60/1.42 oil DIC objective lens.
RIP and RT–qPCR
Approximately 5,000 young adult animals were washed 3 times with PBS–TX (PBS containing 0.01% (v/v) Triton X-100) and crosslinked with 1.8% formaldehyde by rotating at room temperature for 30 min. The reaction was neutralized by adding 125 mM glycine and rotated for 5 min at room temperature. Crosslinked animals were washed with PBS–TX and resuspended in Pierce IP lysis/wash buffer (Thermo Scientific, 1861603) containing 80 U ml–1 RNaseOUT and protease inhibitor cocktail without EDTA (Roche, 11836170001) and sonicated (3 s on, 10 s off, 30% output for 2 min, 2 cycles). Lysates were rotated for an additional 15 min and clarified by centrifuging at 14,000 rpm for 15 min at 4 °C. The concentration of the supernatant was determined using Pierce BCA Protein Assay kits (Thermo Scientific, 23225), and equal amounts of proteins from each sample were used for IP. Anti-Flag M2 magnetic beads (used for 3×Flag::AKAP-17; Millipore Sigma, M8823) were pre-blocked by 1% BSA with rotating 1 h at room temperature and then incubated with supernatants at 4 °C overnight. Beads were washed with Pierce IP lysis buffer for 6 times. Associated RNAs were reverse-crosslinked by incubating at 70 °C for 1 h and extracted with RNA Clean & Concentrator-5 (Zymo Research, R1014). cDNA was synthesized from isolated RNA using SuperScript IV Reverse Transcriptase (Invitrogen, 18090010). Next, 5 ng of input cDNA per reaction was used for analyses. iTaq Universal SYBR Green Supermix (Bio-Rad, 1725120) was used for RT–qPCR analysis. Fold changes were normalized to inputs. All primers used in this study are listed in Supplementary Table 1.
RIP–seq
RIP was performed as described above. To identify potential RNA targets of AKAP-17, approximately 7,000 young adult C. elegans animals expressing 3×Flag::AKAP-17 (tagged group) or AKAP-17 (non-tagged control) were collected for RIP experiments. RIP–seq libraries were prepared following a previously described protocol54, with modifications for C. elegans samples.
In brief, RNA extracted from IP was dephosphorylated using FastAP (Thermo Scientific, EF0651), and cyclic phosphates were removed using T4 polynucleotide kinase (PNK) (NEB, M0201S). RNA was then ligated to an adapter containing a reverse transcription (RT) primer binding site. For input samples, an additional DNase treatment was performed using TURBO DNase (Thermo Scientific, AM2238) to ensure complete removal of gDNA contamination. The RNA was reverse-transcribed into single-stranded cDNA, which was then hydrolysed with NaOH. Following RT, a second adapter was ligated to the ssDNA. PCR amplification was carried out using Illumina-barcoded primers targeting the ligated adapters.
The molarity of PCR-amplified libraries was measured using Agilent TapeStation (G2991BA) DNA ScreenTape (Agilent, 5067–5582). All samples were pooled at equal molarity and subjected to paired-end sequencing (150 bp) on an Illumina NovaSeqX/X+ platforms with 25B flowcells (Genewiz).
Reads were processed using a Snakemake workflow based on the CLAPAnalysis pipeline54, with modified alignment parameters. Paired-end RNA sequencing reads were trimmed to remove adaptor sequences using TrimGalore! (v.0.6.2) and assessed for quality using FastQC (v.0.11.8). Read pairs were first aligned to a combined reference that contained repetitive and structural RNA sequences (ribosomal RNAs, snRNAs, snoRNAs, 45S pre-rRNAs and tRNAs) using Bowtie2 (v.2.5.1). Unaligned reads were subsequently aligned to the C. elegans genome (WBcel235/ce11) using STAR (v.2.7.9a) with the following parameters: –readFilesCommand zcat –alignEndsType Local –outFilterMatchNmin 100 –outFilterScoreMin 100 –outFilterIntronMotifs RemoveNoncanonical –outFilterMultimapNmax 1 –outFilterType BySJout –outSAMunmapped Within –outReadsUnmapped Fastx. PCR duplicates were removed using the MarkDuplicates function in Picard (v.2.18.7).
For each reference gene, we divided the gene body into 100-nucleotide windows and counted the number of reads overlapping each window. Read counts were normalized to the total number of sequenced reads. Gene-level enrichment was calculated by summing the normalized counts across all windows for a gene, and IP enrichment was defined as the ratio of normalized IP to normalized input values. To assess reproducibility between two biological replicates, we defined a reproducibility score based on the difference in enrichment values between IP samples, ranging from 0 (low) to 1 (perfect). Genes with a tag/no-tag fold change greater than 2 and a reproducibility score above 0.55 were defined as AKAP-17-targeted mRNAs. Significance (P values) was assessed using the limma (v.3.62.2) linear modelling framework with an empirical Bayes approach, based on log2-transformed normalized enrichment values. Results are summarized in Supplementary Table 3.
Tc1 excision qPCR assay
Single L4 stage C. elegans animals were transferred onto 6 cm NGM plates and grown at 20 °C until the progeny reached adulthood. After 4 days, worms were collected by washing plates with M9 buffer, and whole-plate lysates were prepared in worm lysis buffer (50 mM KCl, 10 mM Tris-HCl, pH 8.3, and 2.5 mM MgCl2) supplemented with proteinase K (0.2 mg ml–1). Lysates were used directly as DNA templates for qPCR. Reactions were performed with iTaq Universal SYBR Green Supermix (Bio-Rad, 1725121) according to the manufacturer’s instructions, and amplification was carried out on a CFX Connect Real-Time PCR detection system (Bio-Rad, 1855201). Cycling parameters were as follows: initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 61 °C for 30 s (annealing–extension with plate read). eft-3 served as the internal control gene. A nhj-1-null allele (gg875), in which somatic Tc1 excision is suppressed55, was used as a negative control. All primers used in this study are provided in Supplementary Table 1.
CRISPR
C. elegans
All CRISPR deletion or insertion experiments were done using CRISPR-targeted genome-editing techniques56. crRNAs were designed using the IDT online guide RNA design tool (https://www.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE). For oligonucleotide-mediated homology-directed repair (HDR), RNP complexes containing gene-specific crRNA (Supplementary Table 1), tracrRNA (IDT, 1072532) and Alt-R S.p.HiFi Cas9 nuclease V3 (IDT, 1081060) were assembled at 37 °C then mixed with ssODN (IDT, standard desalting; 4 nmol Ultramer) and the co-injection marker PRF4::rol-6(su1006) following manufacturer’s protocols. For dsDNA-mediated HDR (akap-17::mScarlet, caap-1::mScarlet), repair templates that contained 35 bp homologous arms were amplified with PCR, gel-purified and cleaned with 1× to 1.5× AMPure XP reagent (Beckman Coulter, A63881). Diluted repair template (100 ng μl–1) was melted (95 °C 2 min, 85 °C 10 s, 75 °C 10 s, 65 °C 10 s, 55 °C 1 min, 45 °C 30 s, 35 °C 10 s, 25 °C 10 s, 4 °C 10 s, with a 1 °C s–1 ramp down at each step) and mixed with RNPs before microinjection. The injection mix was microinjected into gonads of P0 animals and maintained at 20 °C. Rolling animals, which indicate successful injection, were isolated 4 days later and screened for successful genome deletion or insertion.
For integration of the Tc1::Ngfp SOS splicing reporter (Peft-3::Tc1::unc-54::Ngfp::unc-54 3′ UTR), individual fragments were fused to generate the final genetic constructs using the SapTrap cloning method as previously described57. Engineered transgenes were subcloned into the pDD379 vector and integrated into the genome using CRISPR–Cas9.
Generation of HEK293T knockout cell lines
To generate AKAP17A and CAAP1 knockout cell lines in HEK293T cells, guide RNA (gRNA) sequences targeting AKAP17A or CAAP1 were designed using the CRISPick tool58. cDNA oligonucleotides encoding gRNA sequences (Supplementary Table 1) were synthesized from IDT, annealed and cloned into BsmBI-digested pLentiCRISPR-v2-BFP backbone59,60 using a Quick Ligation kit (NEB, M2200S). HEK293T cells were transfected with plasmids encoding gRNAs using Lipofectamine 3000 transfection reagent (Invitrogen, L3000008). Forty-eight hours after transfection, single BFP-positive cells were isolated by FACS and cultured for 10–14 days for clonal expansion. Clonal populations were screened for knockout validation by immunoblotting, which confirmed the absence of protein expression from targeted genes.
Generation of SOS reporter knock-in cell lines
To generate HEK293T cells expressing Tc1–GFP or HSMAR2–GFP reporters, SOS splicing reporters were cloned into donor plasmid AAVS1-tdTomato targeting vector (Addgene, 194728) using NEBuilder HiFi DNA assembly master mix (NEB, E2621S) with the following modifications: (1) tdTomato was replaced by Tc1–GFP or HSMAR2–GFP reporter cassettes; and (2) the PuroR selection marker was replaced with the BlastR selection marker. The donor plasmids AAVS1-BlastR-Tc1-GFP or AAVS1-BlastR-HSMAR2-GFP, along with a Cas9-expressing plasmid targeting the AAVS1 insertion site (pX458-AAVS1-sg; Addgene, 194721), were co-transfected into HEK293T cells using Lipofectamine 3000 transfection reagent (Invitrogen, L3000008). Three days after transfection, 15 μg ml–1 blasticidin HCl (InvivoGen, ant-bl-05) was added to select for edited cells. After 7 days of selection, surviving cells were collected, and single GFP-positive cells were isolated in a 96-well plate by FACS. Resulting single-cell colonies were expanded, and knock-in cell lines were validated by Sanger sequencing and confirmed by observation of GFP expression.
Lentivirus production and transduction
For lentiviral packaging, HEK293T cells cultured in plain DMEM were transfected with 2.5:1:1.5 ratio of the transfer plasmid pHAGE-2×Flag-AKAK17A, VSV-G envelope-expressing plasmid pMD2.G (Addgene, 12259) and lentiviral packaging psPAX2 (Addgene, 12260) using a CalPhos Mammalian Transfection kit (Takara, 631312). After 12 h, medium was replaced with DMEM containing 10% FBS and the cells were incubated for 48 h to produce lentiviral particles. Virus-containing supernatant was collected at 24 h, 36 h and 48 h after transfection and filtered through a 0.45 μm syringe filter. Lentiviral particles were used to transduce the target cells with 10 μg ml–1 polybrene transfection reagent (Sigma Aldrich, TR-1003-G). Following transduction, cells were selected with 2 μg ml–1 puromycin dihydrochloride (Gibco, A1113803) for 14 days. Puromycin-resistant cells were pooled for downstream analysis.
Flow cytometry
HEK293T cells were rinsed once with PBS and detached from plates using TrypLE Express enzyme (Gibco, 12605028) and passed through a 35 μm nylon mesh strainer (Corning, 352235). Approximately 10,000 individual cells were analysed for B525-FITC-A (green) using a CytoFLEX S flow cytometer. Flow cytometry data were collected using CytExpert software (Beckman Coulter), and figures were created using FlowJo (v.10.7.1) software. A Sony MA900 was used for FACS to isolate isogenic single clones. Flow cytometry gating strategies are provided in Supplementary Fig. 2.
SDS–PAGE and immunoblotting
For cell lysate preparation, HEK293T cells were washed once with ice-cold PBS and lysed on ice for 5 min in RIPA lysis and extraction buffer (Thermo Scientific, 89900) supplemented with PhosSTOP (Roche, 4906845001). Lysates were clarified by centrifugation at 20,000g for 10 min at 4 °C. The supernatants were mixed with 4× SDS sample buffer (Millipore, 70607-3) supplemented with 4% β-mercaptoethanol (Sigma, M6250), denatured at 95 °C for 15 min and resolved using NuPAGE Bis-Tris mini protein gels (Invitrogen, NP0322B0X) in 1× NuPAGE MOPS SDS running buffer (Invitrogen, NP0001).
After SDS–PAGE, proteins were transferred to nitrocellulose membranes (Bio-Rad, 1620112) using a semi-dry transfer method with a Power Blotter station (Invitrogen, PB0010). Membranes were blocked for 1 h at room temperature with Intercept (TBS) blocking buffer (Li-Cor Bio, 927–60001). Primary antibody (Supplementary Table 1) incubation was carried out at 4 °C overnight in TrueBlack WB antibody diluent (Biotium, 23013B-1L). After washing with TBS-T (0.1% Tween 20), membranes were incubated with IRDye secondary antibodies (Li-Cor Bio; Supplementary Table 1) for 1 h. Membranes were washed with TBS-T and imaged using an Odyssey DLx Imaging System (Li-Cor Bio). Uncropped gel images are provided in Supplementary Fig. 1.
IP–MS
A total of 5 × 108 HEK293T cells were seeded 1 day before transfection. HEK293T cells were transfected with plasmid expressing HA-CAAP1 (pGCS-3×HA-CAAP1) using a CalPhos Mammalian transfection kit (Takara, 631312). After 24 h, cells were lysed in 5 ml lysis buffer (40 mM HEPES pH 7.4, 100 mM NaCl and 0.05% CHAPS) with PhosSTOP (Roche, 4906845001) and sonicated as described above. Cell lysates were further rotated for 20 min at 4 °C and cleared by centrifugation at 21,000g for 12 min. Supernatants were incubated with Pierce anti-HA magnetic beads (Thermo Scientific, 88836) at 4 °C for 2 h on a rotator to immunoprecipitate HA-tagged CAAP1 protein complexes. Beads were washed 4 times with lysis buffer, and proteins were eluted using elution buffer (50 mM Tris-HCl pH 7.5, and 10% SDS) by boiling at 95 °C for 4 min.
The eluted proteins were digested with sequencing-grade modified trypsin (Promega, V5113) on S-Trap Micro columns (Protifi, C02-micro-10) according to the manufacturer’s instructions. First, proteins were reduced using 5 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) (Sigma Aldrich, C4706-2G) at 55 °C for 15 min, followed by alkylation with 20 mM iodoacetamide (Sigma Aldrich, I6125) at room temperature in the dark for 30 min. After alkylation, samples were acidified with phosphoric acid (Sigma Aldrich, 345245-100 ml) to a final concentration of 2.5% (v/v). To assist in protein trapping, 10 volumes of 100 mM Tris (pH 7.55) in 90% methanol and 10% water (v/v) were added, and the solution was passed through the S-Trap column by centrifugation at 4,000g for 30 s. Multiple centrifugation rounds were performed to ensure columns were fully loaded. Once proteins were trapped, the column was washed 3 times with 100 mM Tris (pH 7.55) in 90% methanol and 10% water (v/v) and spun dry. Proteins were digested by adding 2 µg trypsin in 20 μl of 50 mM ammonium bicarbonate (pH 8) (Sigma Aldrich, A6141-25G). Digestion occurred overnight at 37 °C in a humidified environment. After digestion, peptides were eluted from the column in three steps, each by centrifugation at 4,000g for 1 min: 40 μl ammonium bicarbonate (pH 8), 40 μl 0.2% formic acid in water and 40 μl 50% acetonitrile (Sigma Aldrich, 34851) in water. Eluted peptides were pooled, dried under reduced pressure using a SpeedVac (Eppendorf, 22820109) and re-suspended in 30 μl of 0.1% formic acid in water. LC–MS/MS data were acquired as previously described61.
A protein database from the Human UniProt SwissProt proteome was used to identify proteins that co-immunoprecipitated with 3×HA–CAAP1. The FragPipe graphical user interface (v.18.0) was used to search data with MSFragger search engine and to post-process results. Tryptic peptides with up to two missed cleavages were included. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine was allowed as a variable modification, with a maximum of four variable modifications per peptide. The allowed mass tolerances were 10 ppm for precursor ions and 0.04 Da for product ions. Peptide hits were filtered to a 1% false discovery rate using PeptideProphet in FragPipe. Peptide counts are provided in Supplementary Table 4.
Co-immunoprecipitation
C. elegans co-IP
Approximately 5,000 young adult animals were collected per sample and resuspended in Pierce IP lysis/wash buffer (Thermo Scientific, 1861603) containing protease inhibitor cocktail without EDTA (Roche, 11836170001) and sonicated using a probe sonicator (10 s on, 10 s off, 50% output for 2 min with probe sonication). Lysates were rotated for 20 min at 4 °C and cleared by centrifugation at 21,000g for 10 min. The protein concentration of supernatant was determined using the BCA method. For each co-IP experiment, equal amounts of proteins from each sample were incubated with Myc-Trap magnetic agarose (ChromoTek, ytma-20) for 2 h at 4 °C. Beads were washed four times with Pierce IP lysis/wash buffer (Thermo Scientific, 1861603). Proteins were eluted by adding 1× SDS sample buffer (Millipore, 70607-3) supplemented with 1% 2-mercaptoethanol (Sigma, M6250), followed by boiling at 95 °C for 15 min. Input and eluted proteins were separated by SDS–PAGE and detected by immunoblotting as described above.
Cell-based co-IP
Co-IP experiments were performed using 2 × 106 HEK293T cells, seeded in 60 mm dishes 1 day before transfection. Cells were transfected with the required plasmids using Lipofectamine 3000 transfection reagent (Invitrogen, L3000008). Forty-eight hours after transfection, whole-cell lysates were prepared by adding 600 μl nuclear lysis buffer (1× PBS, 300 mM NaCl, 1% Triton X-100 and 0.1% Tween 20) supplemented with PhosSTOP (Roche, 4906845001). Lysates were cleared by centrifugation at 20,000g for 10 min at 4 °C and then incubated with anti-Flag M2 magnetic beads (for Flag–AKAP17A; Millipore Sigma, M8823), Pierce anti-HA magnetic beads (for HA–RTCB; Thermo Scientific, 88836). IP was performed by incubating lysates with beads on a tube rotator at 4 °C for 2 h. Beads were washed 3 times with nuclear lysis buffer and once with nuclear dialysis buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl and 0.1% Tween 20). Proteins were eluted by adding 1× SDS sample buffer, followed by boiling at 95 °C for 15 min. Both input and elution fractions were collected and analysed by immunoblotting.
To capture RTCB–CAAP1 and RTCB–AKAP17A interactions, dithiobis (succinimidyl propionate) (DSP) crosslinking was performed as previously described62. Before cell lysis, cells were washed twice with PBS, and protein complexes were crosslinked using 0.1 mM DSP (Thermo Scientific, 22586) in PBS at 37 °C for 30 min. Crosslinking reactions were quenched by adding 20 mM Tris-HCl in PBS (pH 7.4) for 15 min at room temperature. Cells were lysed and proceeded with co-IP protocol described above.
Tripartite split-GFP reporter
To monitor protein–protein interactions for AKAP17A, CAAP1 and RTCB, 5 × 104 HEK293T WT or CAAP1 knockout cells were seeded into 24-well glass-bottom plates (Cellvis, P24-1.5H-N) 1 day before transfection. The next day, cells were co-transfected using using Lipofectamine 3000 transfection reagent with the following encoding plasmids: GFP1–9 (Addgene, 182244), GFP10 (pGCS-HA–GFP10) and GFP11 (pGCS-Flag–GFP11) (negative control); AKAP17A–GFP10, GFP10 and GFP1–9 (negative control for AKAP17A); CAAP1–GFP10, GFP11 and GFP1–9 (negative control for CAAP1); RTCB–GFP11, GFP10 and GFP1–9 (negative control for RTCB); AKAP17A–GFP10, RTCB–GFP11 and GFP1–9 (AKAP17A and RTCB interaction); CAAP1–GFP10, AKAP17A–GFP11 and GFP1–9 (CAAP1 and AKAP17A interaction); CAAP1–GFP10, RTCB–GFP11 and GFP1–9 (CAAP1 and RTCB interaction). Twenty-four hours after transfection, cells were washed with PBS and the culture medium was replaced with live-cell imaging medium. Imaging was performed 30 min after medium exchange using a Nikon Ti2 W1 Yokogawa spinning disk confocal microscope equipped with Plan Apo λD ×60/1.42 oil DIC objective lens.
Illumina sequencing and alternative splicing analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, 15596026) followed by DNase I (Thermo Scientific, EN0521) treatment to remove contaminating DNA. RNA concentrations were determined by Nanodrop 2000, and RNA quality was assessed by gel electrophoresis. rRNA and mtRNA were depleted using 50-nucleotide DNA oligonucleotides complementary to C. elegans rRNA and mtRNA sequences, followed by Thermostable RNase H (NEB, M0523S) treatment, as previously described63. Ribosomal-depleted RNA was then analysed using TapeStation and qPCR, following quality control methods outlined in manufacturer instructions. Libraries were prepared using a KAPA mRNA HyperPrep kit (KR1352-v.4.17) and sequenced with 150-nucleotide paired-end reads, which generated 30 million read pairs on the Illumina NovaSeq 6000 (Biopolymers Facility, Harvard Medical School).
Reads were processed using the Nextflow (v.24.04.4)-based nf-core/rnasplice pipeline64 (v.1.0.4), primarily for downstream rMATS analysis65 (v.4.1.2). Read quality was assessed using FastQC (v.0.12.1), and low-quality reads were trimmed or removed using TrimGalore! (v.0.6.7). Adapter sequences and low-quality bases were clipped. Remaining reads were aligned to the C. elegans genome (WBcel235/ce11) using STAR (v.2.7.9a)66. The resulting alignment was subjected to rMATS for annotation of alternative splicing events. Differential alternative splicing events were identified with the following cut-off criteria: ΔPSI ≥ 0.05 or ≤ −0.05, P value < 0.05. Results were visualized using boxplots.
In vitro protein synthesis and pull-down
HA::RTCB-1 protein was synthesized using a PURExpress In Vitro Protein Synthesis kit (NEB, E6800S) for C. elegans HA::RTCB-1 pull-down experiments. C. elegans rtcb-1 was codon-optimized for E. coli expression and synthesized by Twist Bioscience. The codon-optimized rtcb-1 sequence was cloned into the pDFCI vector with an N-terminal 3×HA tag. Protein synthesis reactions were performed according to manufacturer’s instructions. Following protein synthesis, the reaction was stopped by dilution in 500 μl of Pierce IP lysis/wash buffer (Thermo Scientific, 1861603) and incubated overnight at 4 °C with Pierce anti-HA magnetic beads (Thermo Scientific, 88836) to capture HA::RTCB-1.
For pull-down, worm lysates were prepared as described above and incubated with HA::RTCB-1-bound beads at 4 °C for 2 h. Beads were washed four times with Pierce IP lysis/wash buffer, and bound proteins were eluted using 1× SDS sample buffer. Both input and elution fractions were collected and analysed by immunoblotting.
AlphaFold3 multimer prediction
For each AlphaFold3 protein–protein interaction and docking prediction, the full sequence of proteins was used as input67. AlphaFold3 multimer prediction was performed on the AlphaFold Server (https://alphafoldserver.com) to generate four predicted unrelaxed docking structures with default parameters. For each docking prediction, the highest scoring predicted structure is shown and illustrated using PyMOL (v.2.5.8).
Statistics and reproducibility
For all assays, data are presented as the mean ± s.d. unless otherwise specified in the figure legends. Statistical analyses were performed using GraphPad Prism (v.10). Comparisons between two groups were assessed with unpaired two-tailed Student’s t-tests. For multiple-group comparisons, two-way ANOVA (two-tailed) followed by Tukey’s post hoc test was applied. A P value < 0.05 was considered significant. Exact P values are reported in the figures. All statistical tests were performed on data derived from at least three independent biological replicates. Sample sizes were not predetermined by statistical methods. Randomization was not applied because experiments were designed on the basis of genotype or experimental condition. Investigators were not blinded to allocation; however, control and experimental samples were processed in parallel and handled equally to ensure consistency.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.