Plant material construct development and growth conditions
The background/wild type of osarf1, OE-ARF1, cesa6, arf1cesa6 and NG-CESA6 in this study was rice (Oryza sativa ssp. japonica) variety 9522. Nipponbare (Nip), ein2 and eil1eil2 were sourced from the group of R. Huang. The osarf1, cesa6 and arf1cesa6 knock out alleles were generated using CRISPR–Cas9 technology, with mutant genotyping conducted through PCR amplification of the target regions followed by sequencing of the products. We amplified OsARF1 coding (2,100 bp) and promoter (2,915 bp in front of ATG) sequences and then cloned them into pTCK303 and pCAMBIA1301 to produce OE-ARF1 and ProOsARF1:GUS, respectively. We also amplified OsCESA6 promoter (2,877 bp upstream of ATG) sequence and then cloned the segment into pCAMBIA1301 to produce ProOsCESA6:GUS. mNeonGreen was fused to the OsCESA6 genomic sequence under the control of the OsCESA6 promoter and was cloned into pCAMBIA1301 to create NG-CESA6, as described in reference15. All plasmids were constructed using the In-Fusion HD Cloning Kit (Takara) and were verified by sequencing at BGI, Beijing, China. These plasmids were introduced into the calli of 9522 or cesa6 via Agrobacterium (EHA105)-mediated transformation, with hygromycin B used for selection as outlined in reference32. All primers used are listed in Supplementary Table 1.
All rice samples were grown and collected at the paddy field of Shanghai Jiao Tong University, under natural conditions from June to October. Seedlings were cultivated in a plant incubator at 28 °C with a 16 h:8 h light:dark cycle and 50%–70% humidity.
Soil compaction details
Loamy sand from the Newport series (comprising 83.2% sand, 12.1% clay and 4.7% silt; with 2.93% organic matter and a pH of 6.35; classified as FAO Brown Soil) and clay soil were collected from the University of Nottingham farm at Bunny, Nottinghamshire, UK (52.52° N, 1.07° W). Both soils were passed through a 2-mm sieve. The moisture content was determined by drying the soil at 45 °C until a constant weight was achieved.
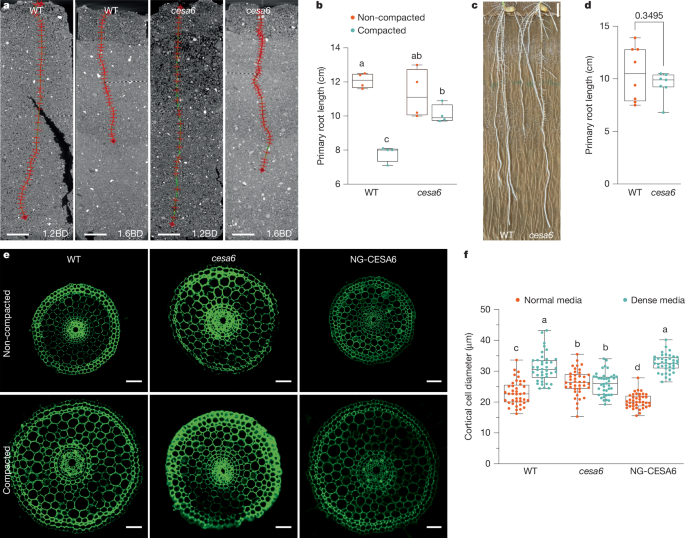
Mesocosms were prepared by packing them with the sieved soil to achieve a bulk density of 1.2 g cm−3, to simulate a non-compacted condition, and 1.6 g cm−3 for a compacted condition. Rice seeds were sterilized with a 25% bleach solution for 10 min and subsequently rinsed six times with sterilized water. The seeds were then placed on filter paper for 48 h in a dark incubator chamber to allow for germination. One germinated seedling per mesocosm was positioned on the soil surface and covered with a 1 cm top layer of soil at 1.2 g cm−3, in both non-compacted and compacted soil conditions. The seedlings were then placed in a rice growth chamber, which was maintained at 28 °C, with a 12 h photoperiod and 70% relative humidity.
X-ray CT imaging
Five-day-old seedlings of wild type, osarf1 and cesa6 were grown in 3D-printed columns (33 mm in diameter and 100 nm in height) filled with sandy loam soil under non-compacted (1.2 g cm−3) and compacted (1.6 g cm−3) conditions, respectively. They were then imaged using a GE Phoenix v|tome|x M 240 kV X-ray tomography system at The Hounsfield Facility, University of Nottingham. Three-dimensional image reconstruction was carried out using Datos|REC software (GE Inspection Technologies). The roots were segmented from the soil using a polyline tool in VGStudioMAX V2.2 (Volume Graphics) to demonstrate the root length phenotype. The scanning protocol involved collecting 3,240 projection images in FAST mode (continuous rotation), with the X-ray tube set to an energy of 140 kV and a current of 200 μA. The detector’s exposure time was 131 ms, and the voxel resolution was 57 μm. Each scan had a duration of 7 min.
Agar experimental details
Agar at concentrations of normal (0.3%) and dense (0.6%) (Sigma Aldrich 7002) was boiled and poured into tanks measuring 7.5 cm in diameter and 10 cm in length to simulate non-compacted and compacted soil conditions, respectively. After the agar solidified, germinated seeds were placed on the media surface, followed by watering (1 cm height). The seeds were then grown in a plant incubator (28 °C, 16 h light:8 h dark, 70% relative humidity) for 5 days. Finally, root phenotypes were imaged using a Nikon camera, and the images were analysed with ImageJ software.
Imaging of root tip thickness
Root tips, including the root cap, meristematic, elongation and differentiation zones (approximately 1 cm of the rice root tip), were washed 3 times with sterilized water. They were then embedded in 5% melted agarose (LabTop Biotechnology). Transverse sections, 50 μm thick, were cut using a Leica Vibratome (VT 1000 S) and imaged with a Leica SP5 confocal microscope utilizing the UV laser.
RNA isolation and RT–qPCR
Root tips, including the root cap, meristematic zone, elongation zone, and differentiation zones (approximately 1.5 cm of the rice root tip), grown in soil or agar media conditions, were sampled in three biological replicates. Total RNA was extracted using Trizol reagent (Invitrogen) followed by purification with chloroform/isopentyl alcohol. The cDNA was synthesized using the FastQuant RT Kit with gDNase (Tiangen Biotech). RT–qPCR was conducted using the SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology). The reaction mixture consisted of 7.5 μl SYBR, 2 μl cDNA, 0.3 μl each of forward and reverse primers, and 4.9 μl double-distilled water. The Ubiquitin gene served as the reference for assessing gene expression levels. All RT–qPCR primers are listed in Supplementary Table 1, and gene information is listed in Supplementary Table 2.
Y1H screening
Nine promoter fragments—753 bp for OsCESA1, 1,973 bp for OsCESA3, 1,928 bp for OsCESA5, 1,964 bp for OsCESA6, 1,751 bp for OsCESA8, 1,943 bp for OsCSI1, 1,872 bp for OsCSLF6, 2,628 bp for OsCTL, and 923 bp for OsSTL—as well as a 1,738 bp fragment for the OsHARPIN1 promoter were cloned into the R4L1pDEST_HIS2 plasmid33. The cDNA library encompasses 1,143 rice transcription factors, representing all known transcription factor families. The appropriate 3AT concentration was determined through autoactivation testing. Subsequently, plasmids were transformed into YM4271 yeast strain (Clontech/TAKARA) using TE/LiAc and PEG solution. Yeast plates were incubated at 30 °C for 3–7 days, after which images were captured using a camera.
Dual-luciferase assay
Rice seedlings were grown for 7 days post-germination in a plant incubator set to 28 °C with a 16 h:8 h light:dark cycle and 50%–70% humidity. Seedling shoots were collected and digested in an enzyme solution containing 0.2 g Cellulase ‘Onozuka’ RS (Yakult), 0.05 g Macerozyme R10 (Yakult) and β-mercaptoethanol to a final concentration of 10 mM. After degassing twice, the solution was incubated at 28 °C and 80 rpm for 2 h. Protoplasts were isolated using W5 solution (150 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7) and MMg solution (400 mM mannitol, 15 mM MgCl2·6H2O, 4 mM MES, pH 5.7), examined under a microscope, and adjusted to the appropriate concentration (2.0–2.5 × 105 cells per ml). The OsARF1 and eGFP coding sequences, serving as effectors, were cloned into the pZmUBQ1p_SX_HSPG plasmid. Similarly, promoter sequences of cellulose synthesis genes, acting as reporters, were cloned into the pGL4.1HSP plasmid. Both effectors and reporters were introduced into the protoplasts using PEG/Ca2+ and incubated for 16–18 h at 28 °C with 80 rpm agitation. The relative reporter activity was determined by normalizing the reporter luciferase value to the Renilla luciferase reference value. The incubated protoplasts were lysed using the Dual-LUC reporter assay kit (TOYOINK) and the luminescence was measured using a multi-scan spectrometer (TECAN).
EMSA
First, we amplified the full-length coding sequence of OsARF1 and cloned it into the pSP6-T7 vector (Promega). The OsARF1 protein was then expressed using an in vitro translation kit (TNT T7/SP6 Coupled Wheat Germ Extract System; Promega) and analysed by western blot (anti-His: M20003, Abmart, 1:2,000 dilution; Goat Anti-Mouse IgG HRP: M21001, Abmart, 1:5,000 dilution). Double-stranded M1, M2 and M3 hot probes were labelled with FAM dye at the 5′ end. For competition assays, we used 200-fold excess of non-labelled probes (wt) and labelled mutated probes (mut). The protein-probe binding reaction mixture, which included 250 mM Tris-Acetate, 10 mM DTT, 1 mg ml−1 BSA, and 20 mM magnesium acetate, was incubated at 25 °C for 20 min. The reaction products were then resolved on a 6% native polyacrylamide gel and visualized using the Cy2 channel of a ChemiDoc MP imaging system (Bio-Rad).
ChIP–qPCR
The ChIP assay was conducted as described34, with slight modifications. Approximately 2 g of root tips (about 1.5 cm in length) from wild-type and ProOsARF1:gOsARF1–GFP plants (verified by anti-GFP: G1544, Sigma, 1:5,000 dilution; and Goat Anti-Rabbit IgG Antibody: AP132, Sigma, 1:5,000 dilution) were crosslinked with 1% (v/v) formaldehyde in extraction buffer (0.4 M sucrose, 10 mM Tris-HCl, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and a protease inhibitor cocktail, pH 8.0), then pulverized in liquid nitrogen. Chromatin was subsequently isolated and sonicated to generate DNA fragments ranging in size from 200 to 500 bp. GFP-Trap Magnetic Agarose (ChromoTek) was utilized to precipitate OsARF1–DNA complexes. Fragments of the OsCESA5 and OsCESA6 promoters were quantified by RT–qPCR using primers listed in Supplemental Table 1. Enrichment in the ProOsARF1:gOsARF1–GFP samples was compared with the levels in wild-type plants.
Hormone and indaziflam treatment conditions
Rice seeds of ProOsARF1:GUS were sterilized with 70% (v/v) ethanol for 1 min and 3% (v/v) sodium hypochlorite solution for 15 min, followed by at least 6 rinses with sterile double-distilled water, for 3 min each. The seeds were then placed on filter paper in germination boxes for 3 days under a 16 h:8 h light:dark cycle. After germination, the seeds were exposed to various conditions: mock (water), 1 μM IAA, 25 μM ABA and 100 μM ACC for 2 days.
Similarly, germinated seeds of wild-type, osarf1 and OE-ARF1 plants were transferred to black boxes containing growth medium supplemented with either 150 pM (final concentration) DMSO or 150 pM indaziflam, and 250 pM DMSO or 250 pM indaziflam, and 50 nM DMSO or 50 nM DCB. These were incubated in a plant incubator at 28 °C, with a 16 h:8 h light:dark cycle and 50%–70% humidity for 5 days. The primary root lengths of the mock-treated and experimental groups were measured and compared using ImageJ. This experiment was performed in triplicate.
GUS histochemical and quantification
Seedlings of the rice auxin response factor transcriptional reporter (ProOsARF1:GUS and ProOsCESA6:GUS) were grown in conditions including mock (water with added DMSO), 0.6% agar media, 1 μM IAA, 25 μM ABA and 100 μM ACC. Root tips were then collected and incubated overnight in GUS buffer (50 mM NaPO4 buffer, pH 7.0, 10 mg ml−1 X-Gluc, and 0.02% (v/v) Triton X-100) at 37 °C. Subsequently, the samples were washed with 70% (v/v) ethanol until they became transparent. Cross-sections were prepared using a Leica vibratome (VT 1000 S) at a thickness of 50 μm. Images were taken with a Leica light microscope (M205A) equipped with a CCD camera, and additional photographs were obtained using a Nikon H600L light microscope (Tokyo).
For GUS activity quantification, approximately 0.1 g of ProOsARF1:GUS roots (1.5 cm) were collected after 2-day treatment with mock (DMSO), 1 μM IAA, 25 μM ABA, or 100 μM ACC. Root tissues were ground in liquid nitrogen and homogenized in 1 ml GUS extraction buffer (1 M Na2HPO4, 1 M NaH2PO4, 10% SDS, 0.5 M EDTA, Triton X-100, β-mercaptoethanol). The homogenate was centrifuged at 12,000 rpm for 5 min at 4 °C, and protein concentration in the supernatant was determined using the Bradford method. For the GUS activity assay, 100 μl of protein extract was mixed with 400 μl pre-warmed (37 °C) GUS extraction buffer and 500 μl MUG substrate (2 mM). The reaction mixture was incubated at 37 °C, and 200 μl aliquots were collected at 0, 15, 30, 45 and 60 min intervals. Each aliquot was immediately mixed with 800 μl stop solution (0.2 M Na2CO3) and stored at room temperature in the dark. Fluorescence intensity was measured using a fluorescence microplate reader (excitation: 365 nm, emission: 455 nm, slit width: 10 nm). The rate of fluorescence intensity change was calculated from the slope of the fluorescence intensity versus time plot and normalized to total protein content to determine specific GUS activity.
Cellulose measurements
Cellulose content was quantified as described35, with modifications. Five-day-old seedlings of wild-type, osarf1 and OE-ARF1 were grown in 0.3% or 0.6% agar media (Sigma-Aldrich-7002) at 28 °C under a 16 h:8 h light:dark cycle. Root tips (approximately 1.5 cm) were collected and processed to obtain alcohol-insoluble residue (AIR). In brief, samples were incubated in 1.5 ml 70% ethanol at 70 °C for 1 h, followed by a second 45 min incubation with fresh ethanol. After ethanol removal, samples were treated with 1 ml acetone at room temperature for 2 min. The resulting AIR was dried, weighed, and transferred to 15 ml Falcon tubes. For cellulose extraction, 3 ml of acetic/nitric reagent was added to each sample (including a blank control), and tubes were heated in a boiling water bath for 30 min. After cooling, samples were centrifuged at 2,000g for 10 min. The supernatant was carefully discarded, and pellets were washed with 8 ml water, gently resuspended and incubated for 15 min before centrifugation (2,000g, 10 min). A final wash was performed with 4 ml acetone, followed by a 5 min incubation and centrifugation (2,000g, 10 min). Samples were dried in a vacuum oven at 30–40 °C. The dried samples were hydrolysed in 1 ml of 67% sulfuric acid with shaking (180 rpm) at room temperature until complete dissolution. For colorimetric determination, 40 μl of each hydrolysate was mixed with 1 ml ice-cold anthrone reagent and heated in boiling water for 5 min, followed by immediate ice quenching. Cellulose content was calculated as percentage of cell wall (CW) per AIR using glucose standard curves according to the formula: CW (%) = (glucose (μg))/(AIR (μg)) × 100% × (H2SO4 volume)/(sample volume).
Cell wall thickness experimental details
Roots from 5-day-old seedlings of wild-type, osarf1, OE-ARF1 and cesa6 plants were cultivated in 0.3% and 0.6% agar media (Sigma-Aldrich-7002) at 28 °C under a 16 h:8 h light:dark cycle. Root tips, approximately 1 cm in length, were collected and fixed in a solution of 3% (w/v) formaldehyde (formaldehyde freshly prepared from paraformaldehyde) and 0.25% glutaraldehyde in 0.2 N sodium phosphate buffer (pH 7.0). These root tips were then post-fixed in 2% OsO4 in PBS (pH 7.2). The samples were dehydrated through a graded ethanol series (70% for 30 min, 90% for 30 min, and 100% 3 times, each for 30 min) and then transitioned through ethanol/epoxypropane mixtures (2:1, 1:1 and 1:2), followed by pure epoxypropane, each for 10 min. Samples were embedded in acrylic resin (London Resin Company) and placed in a drying oven at 37 °C for 5–12 h to eliminate bubbles, then the temperature was increased to 45 °C for 2 h, and finally to 65 °C for 48 h. We carefully selected comparable elongation zone regions among all genotypes by 2-µm semi-thin sections (Extended Data Fig. 8a), and then made ultra-thin sections (50 to 70 nm; Extended Data Fig. 8b), which were double-stained with 2% (w/v) uranyl acetate and 2.6% (w/v) lead citrate aqueous solution and examined with a JEM-1230 transmission electron microscope (JEOL) at 80 kV. Wall thickness was estimated via ImageJ software. To account for cell wall thickness heterogeneity, we measured a single wild-type cortical cell divided into 8 segments and found that cell wall thickness variations can effectively be averaged by measuring wall thickness at 15 different points (Extended Data Fig. 9). Therefore, we selected 15 points around each cell of a total of 20 cells per genotype. These experiments were repeated five times.
Cell wall stiffness experimental details
Roots from 5-day-old seedlings of wild-type, osarf1, OE-ARF1, cesa6 and arf1cesa6 plants were grown in 0.6% agar media (Sigma-Aldrich-7002) at 28 °C with a 16 h:8 h light:dark cycle. Primary root tips, measured to 1.5 cm, were embedded in 5% melted agarose (LabTop Biotechnology) for sectioning. Longitudinal sections of 50 µm thickness were produced using a Leica vibratome (frequency 50 Hz, amplitude 1 mm, VT 1000 S), and light microscopy was employed to ensure the stele and cortical tissues were correctly positioned with the elongation zone visible. The sections were then stored in deionized water at 4 °C overnight.
AFM assays were conducted following the methods described36. A Dimension ICON (Bruker Nano) equipped with NanoScope Analysis 9.4 software was utilized to probe all root samples. The MLCT-C probe (Bruker Nano), with an average spring constant of 0.01 N m−1, indentation depth (varying from samples, 200–300 nm), speed (3,000 nm s−1), and force limit (300 pN) was used in the experiments. Root sections set in agarose were affixed to glass slides with tape and hydrated with deionized water for 30 min prior to AFM analysis. While operating in force-spectroscopy mode under water-hydrated conditions, 10–15 independent areas within the visible cortex and epidermis zones (as shown in Fig. 4a) were examined. Four biological replicates were conducted for each plant line. Apparent stiffness (in pN nm−1) values were extracted from individual force-distance curves using a contact point based fit and a linear stiffness model, using NanoScope Analysis 1.8 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.