Plant materials and growth conditions
The Arabidopsis thaliana Columbia (Col-0) ecotype was used as the main experimental organism. Seeds of Col-0 (N70000), fd-3 (SALK_054421), ft-10 (GK_290E08) and other transgenic plants were surface-sterilized with 70% ethanol for 10 min, rinsed with 99% ethanol for 5 min, air-dried and stratified at 4 °C for 3 days before sowing. Plants were grown on soil under long-day conditions (16 h light:8 h dark cycles) or were grown vertically on plates containing 1% agar supplemented with half-strength Murashige and Skoog (MS) medium (pH 5.7) at 22 °C with a light intensity of 160–180 μmol m−2 s−1 provided by LED bulbs (Philips F17T8/TL841 17 W).
Plasmid construction
To generate different epitope-tagged fusions of FD, the genomic fragment carrying FD promoter (2,930 bp), the full-length coding region and the 3′ untranslated region (1,982 bp) were amplified from Col-0 genomic DNA. DNA encoding 3HA-3Flag and 2HA-mVenus57 tags were synthesized and amplified with PrimeSTAR GXL DNA Polymerase (Takara Bio) for subcloning. Overlapping PCR was then performed to obtain genomic fusions with FD and the epitope tags. Corresponding PCR fragments were then cloned into a modified binary vector PER8 using a HiFi DNA Assembly Kit (NEB). The constructs were transformed into fd-3 mutants using the floral dip method. The same strategy (with genomic fusions) was used to construct gGRFs::2HAmScarlet-I58-GRFs in Col-0 (GRF2 and GRF6), grf7 (SALK_084141)or grf8 (SALK_148929) mutants, and three genomic constructs carrying the 9,149-bp promoter, the FT coding region fused with different tags (gFT::FT-mVenus, gFT::FT-Venus-Halo-Venus and gFT::FT-ALFA59), and a 3,159-bp downstream sequence were transformed separately into ft-10 (GK-290E08) plants. SUC2::pp2A.1-mCherry was transformed in to Col-0 background. Primers used to amplify these sequences are listed in Supplementary Table 4.
Generation of Arabidopsis mutants and transgenic plants
Corresponding Col-0, fd-3 or ft-10 mutant plants were grown in the greenhouse under LD conditions and were transformed by the floral dip method using Agrobacterium tumefaciens strain GV3101. The resulting transgenic T1 seeds were screened on half-strength MS medium with supplemented hygromycin for 7 LD and then they were transferred to soil for the measurement of flowering time.
RNA extraction and RT–qPCR analysis
Total RNA was extracted from 13-day-old seedlings grown in long-day conditions using the RNeasy plant Mini Kit (QIAGEN) with an on-column DNase (QIAGEN) treatment. cDNA was synthesized from 1 μg RNA using a QuantiTect Reverse Transcription Kit (QIAGEN). Real-time PCR was performed with iQ SYBR Green Supermix (Bio-Rad) in a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The reference gene ACTIN2 was used for normalization. Three technical replicates for each of three independent biological replicates were performed for each experiment and representative results are presented. The primers used for qRT–PCR are listed in Supplementary Table 4.
Immunoblot assays
For western blots, approximately 30 mg of tissue from 13-day-old seedlings grown in long-day conditions was ground into fine powder with liquid nitrogen with a TissueLyser system (QIAGEN). Total protein was extracted using denaturing buffer (100 mM Tris-HCl pH 7.5, 100 mM NaCl, 30 mM EDTA pH 8.0, 4% (w/v) SDS, 20% (v/v) glycerol, 20 mM β-mercaptoethanol (Sigma-Aldrich), 20 mM DTT, 2 mM PMSF (Sigma-Aldrich), 1× Protease Inhibitor Cocktail (PIC, Sigma-Aldrich, P9599), 1× Phosphatase Inhibitor Cocktail 2 (PIC2, Sigma-Aldrich, P5726), 1× Phosphatase Inhibitor Cocktail 3 (PIC3, Sigma-Aldrich, P0044), 80 μM MG132 (Sigma-Aldrich), and 0.01% bromophenol blue) in a 1:5 (w/v) ratio and was boiled at 95 °C for 10 min. Protein samples were centrifuged at 16,000g for 5 min at room temperature and the supernatants were transferred to a new low-protein-binding tube and separated by SDS–PAGE.
For immunoblotting, separated proteins from the gels were transferred onto a PVDF membrane by the Trans-Blot Turbo Transfer System (Bio-Rad). Blots were probed with anti-H–horseradish peroxidase (HRP) (Roche, 12013819001, 1,000-fold dilution), anti-ALFA–HRP (NanoTag, N1505, 1:2,000-fold dilution) or anti-actin–HRP (Santa Cruz, sc-47778, 1:5,000-fold dilution) antibodies conjugated to HRP were used at 1:2,000-fold dilution, in TBS-T buffer. The blots were developed with a 1:1 mix of SuperSignal West Femto Maximum Sensitivity and SuperSignal West Dura Extended Duration Substrates and signals were detected on a ChemiDoc MP Imaging System (Bio-Rad). Uncropped blots are shown in Supplementary Fig. 4.
Co-immunoprecipitation assay
The in vivo co-immunoprecipitation assays were performed as previously described, with minor modifications60. In brief, 3 g of material from 12-day-old seedlings grown in long-day conditions was collected at Zeitgeber time (ZT) 7 and cross-linked in 1× phosphate-buffered saline (PBS) with 1 mM disuccinimidyl glutarate with vacuum filtration for 15 min. The tissues were washed and frozen in liquid nitrogen before storing at −80 °C. The tissues were ground to a fine powder in liquid nitrogen, and semi-pure nuclei were extracted in nuclei isolation buffer (10 mM Tris-HCl pH 8.0, 400 mM sucrose, 0.05% Triton X-100, 1 mM PMSF, 5 mM β-mercaptoethanol and 0.25× protease inhibitor cocktail (PIC)). The isolated nuclei were washed 3 times in wash buffer (10 mM Tris-HCl pH 8.0, 250 mM sucrose, 0.5% Triton X-100, 10 mM MgCl2, 1 mM PMSF, 5 mM β-mercaptoethanol and 0.25× PIC). Nuclear proteins were released by brief sonication in the buffer (Tris-HCl pH 7.5, 3 mM EDTA, 0.5% Triton X-100, 150 mM NaCl, 1 mM PMSF, 50 μM MG132, 1 mM DTT, 1× PIC, 1× PIC2 and 1× PIC3). Western blotting was performed with the extracted nuclear proteins using anti-HA (12013819001, Roche) and anti-H3 (Abcam, ab1791) antibodies before immunoprecipitation.
For co-immunoprecipitation, 30 μl anti-HA magnetic beads (Thermo Fisher) was added to the diluted nuclear protein solution (0.5% Triton X-100, 1 mM EDTA, 20 mM Tris-HCl pH 7.5, and 100 mM NaCl and 1× PIC (Sigma-Aldrich)) and rotated for 40 min at 4 °C. The beads were washed five times with immunoprecipitation buffer. Aliquots (3 μl) of beads were boiled with 2× Laemmli buffer (Bio-Rad) for immunoblotting analysis using anti-HA (Roche, 12013819001, 1:1,000-fold dilution), anti-Strep (IBA, 2-1509-001, 1:4,000-fold dilution) and anti-H3 (Abcam, ab1791, 1:4,000 dilution). The remaining beads with the IPed proteins were stored at −80 °C before on-bead digestion for liquid chromatography–mass spectrometry.
Mass spectrometry and data analysis
The anti-HA magnetic beads with IPed proteins were digested on beads using trypsin or LysC to identify interacting proteins of FD, GRF7 or GRF8, or phosphorylation peptides of FD. In brief, the beads were buffer exchanged and re-dissolved in 25 µl digestion buffer I (50 mM Tris pH 7.5, 2 M urea, 1 mM DTT, 5 ng µl−1 trypsin) and incubated for 30 min at 30 °C in a Thermomixer at 400 rpm. Next, beads were pelleted, and the supernatant was transferred to a new tube. Digestion buffer II (50 mM Tris pH 7.5, 2 M urea, 5 mM chloroacetamide) was added to the beads and after mixing the beads were pelleted, the supernatant was collected and combined with the previous one. The combined supernatants were then incubated overnight at 32 °C in a Thermomixer at 400 rpm; samples were protected from light during incubation. The digestion was quenched by adding 1 µl trifluoroacetic acid (TFA) and desalted with C18 Empore disk membranes according to the StageTip protocol61.
Dried peptides were re-dissolved in 2% acetonitrile (ACN), 0.1% TFA (10 µl) for analysis. Samples were analysed using an EASY-nLC 1200 (Thermo Fisher) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher). Peptides were separated on 16-cm frit-less silica emitters (New Objective, 75-µm inner diameter), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9 µm resin (Dr. Maisch). Peptides were loaded onto the column and eluted for 115 min using a segmented linear gradient of 5% to 95% solvent B (0 min: 5% B; 0–5 min 5% B; 5–65 min →20% B; 65–90 min →35% B; 90–100 min →55% B; 100–105 min →95% B; 105–115 min 95% B) (solvent A: 0% ACN, 0.1% formic acid; solvent B: 80% ACN, 0.1% formic acid, solvents A and B together constituting 100% of the mobile phase) at a flow rate of 300 nl min−1. Mass spectra were acquired in data-dependent acquisition mode with a TOP15 method. MS spectra were acquired in the Orbitrap analyser with a mass range of 300–1,750 m/z at a resolution of 70,000 full width at half maximum (FWHM) and a target value of 3 × 106 ions. Precursors were selected with an isolation window of 1.3 m/z (Q Exactive Plus). HCD fragmentation was performed at a normalized collision energy of 25. MS/MS spectra were acquired with a target value of 105 ions at a resolution of 17,500 FWHM, a maximum injection time of 55 ms and a fixed first mass of m/z 100. Peptides with a charge of +1, greater than 6, or with unassigned charge state were excluded from fragmentation for MS2, and dynamic exclusion for 30 s prevented repeated selection of precursors.
Alternatively, samples were analysed using an Ultimate 3000 RSLC nano (Thermo Fisher) coupled to an Orbitrap Exploris 480 mass spectrometer equipped with a FAIMS Pro interface for Field asymmetric ion mobility separation (Thermo Fisher). Peptides were pre-concentrated on an Acclaim PepMap 100 pre-column (75 µM × 2 cm, C18, 3 µM or 5 µM, 100 Å, Thermo Fisher) using the loading pump and buffer A (water, 0.1% TFA) with a flow of 7 µl min−1 (3 µM), or 15 µl min−1 (5 µM) for 5 min. Peptides were separated on 16-cm frit-less silica emitters (New Objective, 75 µm inner diameter), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9 µm resin (Dr. Maisch). Peptides were loaded onto the column and eluted for 130 min using a segmented linear gradient of 5% to 95% solvent B (0 min: 5% B; 0–5 min 5% B; 5–65 min →20% B; 65–90 min →35% B; 90–100 min →55% B; 100–105 min →95% B; 105–115 min 95% B; 115–115.1 min →5% B, 115.1–130 min 5% B) at a flow rate of 300 nl min−1. Mass spectra were acquired in data-dependent acquisition mode with the TOP_S method using a cycle time of 2 s. For field asymmetric ion mobility separation (FAIMS), two compensation voltages (−45 and −60) were applied and the cycle time was set to 1 s for each experiment. MS spectra were acquired in the Orbitrap analyser with a mass range of 320–1,200 m/z at a resolution of 60,000 FWHM and a normalized AGC target of 300%. Precursors were filtered using the MIPS option (MIPS mode = peptide), the intensity threshold was set to 5,000, Precursors were selected with an isolation window of 1.6 m/z. HCD fragmentation was performed at a normalized collision energy of 30%. MS/MS spectra were acquired with a target value of 75% ions at a resolution of 15,000 FWHM, inject time set to auto, and a fixed first mass of m/z 120. Peptides with a charge of +1, greater than 6, or with unassigned charge state were excluded from fragmentation for MS2.
Raw data were processed using MaxQuant software62 (v.1.6.3.4, http://www.maxquant.org/) with label-free quantification (LFQ) and iBAQ enabled63. MS/MS spectra were searched using the Andromeda search engine against a combined database containing the sequences from A. thaliana (TAIR10_pep_20101214; ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/) and sequences of 248 common contaminant proteins and decoy sequences. Trypsin or LysC specificity was required and a maximum of two missed cleavages allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of cysteine residues was set as fixed, and oxidation of methionine and protein N-terminal acetylation were set as variable modifications. Peptide-spectrum matches and proteins were retained if they were below a false discovery rate (FDR) of 1%.
For interacting protein analyses, statistical analysis of the MaxLFQ values was carried out using Perseus (v.1.5.8.5, http://www.maxquant.org/). Quantified proteins were filtered for reverse hits and hits ‘identified by site’ and MaxLFQ values were log2-transformed. After grouping samples by condition, only those proteins were retained for the subsequent analysis that had two valid values in one of the conditions. Two-sample t-tests were performed using a permutation-based FDR of 5%. Alternatively, quantified proteins were grouped by condition and only those hits were retained that had three valid values in one of the conditions. Missing values were imputed from a normal distribution (1.8 downshift, separately for each column). Volcano plots were generated in Perseus using an FDR of 5% and an S0 = 1. The Perseus output was exported and further processed using Excel.
To identify phosphorylation of T282, the Phospho (STY)Sites.txt file was manually inspected for the presence and localization of the site. The presence of the site was confirmed by searching individual raw files using ProteomeDiscoverer 2.2 (Thermo Fisher).
Confocal imaging of SAM cells
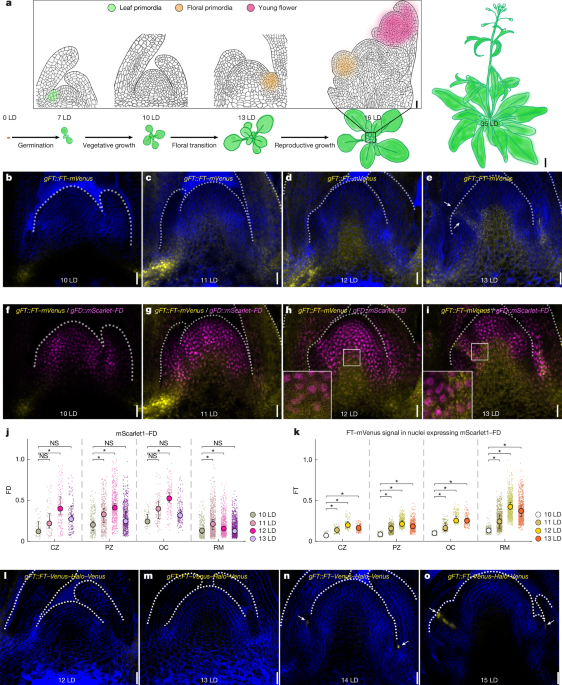
SAMs of seedlings grown in long-day conditions were dissected and fixed with 4% (w/v) paraformaldehyde. The fixed samples were washed twice with PBS for 5 min and cleared with ClearSee solution64 for 2 days in the dark at room temperature. After clearing, samples were washed twice with PBS buffer for 5 min and embedded with 6.5% (w/v) low-melt agarose (Bio-Rad). The embedded samples were sectioned into 70-μm slices using a vibrating blade microtome (Leica VT1000 S) and then stained with dyes. For the FD and GRF co-localization analysis, the cell wall was stained with Renaissance 2200 (0.1% (v/v) in PBS)65 for 30 min and washed in PBS buffer for 5 min. For the FD-chromatin co-localization analysis, the cell wall was stained with Direct Red 23 (0.5% (w/v) in ClearSee)66 for 1 h and washed with PBS buffer for 10 min. The nuclear chromatin was then stained with 1 μg ml−1 DAPI (Thermo Fisher) for 30 min and washed in PBS buffer for 10 min. The stained samples were mounted onto slides with ProLong Antifade Mountants (Thermo Fisher) for signal preservation. Image collection was performed using a Zeiss LSM 880 confocal microscope. The Renaissance and DAPI signals were detected at 410–503 nm with an excitation wavelength of 405 nm. The mVenus signal was excited with a 514 nm laser and collected at 520–560 nm and mScarlet-I and Direct Red 23 were excited with a 561 nm laser and detected at 566–620 nm. The imaging data were processed using Zen 3.10 (Zeiss) software.
Single-cell nuclear quantification of FD–FT fluorescence signal
Individual confocal images of gFT::FT–mVenus and gFD::mScarlet1–FD were processed using Cellpose (2.2.3)67,68 and Matlab (MathWorks (2022); MATLAB v.9.13.0 (R2022b)). Nuclear segmentation was performed using cyto Cellpose model on the FD-channel.tif file. The cell diameter parameter was automatically calibrated. The output nuclear segmentation.png files were processed using custom-made MATLAB code and adapting the previous method68 to 2D images. For each confocal image, a curved line was drawn following the parabolic outline of the SAM. A parabolic fit was then performed, accounting for a possible tilt of the SAM. Based on the fitted parabola, a 2D parabolic mask was created. A rectangular mask was also created, extending from the two ends of the parabola up to the inferior edge of the image. These two masks were combined and all intensity values of the pixels outside the new mask were set to 0. Using the previously generated parabolic mask and published WUS/CLV3 data68, the meristematic tissue was divided into four different regions: central zone (CZ), organizing centre (OC), peripheral zone (PZ) and rib meristem (RM). The first two regions were defined using the height and width of CLV3 and WUS domains, respectively, as proxies. All meristematic tissue below the OC and within the previously generated rectangular mask was considered to be RM. PZ included all nuclei located in the region contained between the OC/CZ and the fitted parabola. Such parametrization of the SAM allows for its compartmentalization it into four different regions and for the assignment of each nucleus to its associated regions if its centroid coordinates are contained that region. Brunner–Munzel test was used to measure statistic significant differences in the median distributions of FD and FT nuclear concentrations between time points. Further details are at https://gitlab.com/GRM_14/gao_ding_et_al_2025/-/tree/011d3d70fc1c0f41670c6f0b860b3c586c8949fd/.
RNA in situ hybridization
RNA in situ hybridization was performed as described previously69. The template for the FT probe was transcribed from cDNA using a specific primer pair (Supplementary Table 4) with T3 and T7 polymerase binding sites attached to the forward and reverse primers, respectively.
RNAscope fluorescent multiplex assays
The RNAscope assay was conducted following the RNAscope Multiplex Fluorescent Assay v.2 protocol provided by ACDBio (materials available at https://acdbio.com). In brief, formalin-fixed, paraffin-embedded tissue samples were used for analyses. Specific probes for FD, FT and TFL1 (assigned to channels C1, C2 and C3, respectively) with the following catalogue numbers: 1307011-C1 (FD), 1307021-C2 (FT), and 1307031-C3 (TFL1) were used. To visualize FD, FT and AP1 in the same sample, the AP1 probe was assigned to channel C3 (1569941-C3). The RNAscope 3-plex Negative Control Probe (320871) was used as a negative control. All probes were hybridized overnight at 40 °C.
To visualize targets, TSA Plus fluorophores (diluted with TSA buffer from ACDBio) were applied as follows: TSA Vivid 520 (323271, diluted 1:2,500) for C3, TSA Vivid 570 (323272, diluted 1:1,500) for C1, and TSA Vivid 650 (323273, diluted 1:1,500) for C2. Additionally, Renaissance (0.1% v/v in PBS) was used to stain the cell walls.
Confocal images were captured using a Zeiss LSM 880 confocal microscope. The imaging data were processed and analysed using Zen 3.10 (Zeiss), Fiji (v.2.16.0), Cellpose (v.2.2.3) and Matlab (v.9.13.0 R2022b). The Renaissance signal was detected at 410–503 nm with an excitation wavelength of 405 nm. The filter settings for FITC, Cy3 and Cy5 were used for the TSA Vivid Fluorophore 520, 570 and 650, separately.
Quantification of RNAscope images
Quantification of RNAscope images at the tissue level was performed using a similar pipeline as the custom-made MATLAB described before, except that first, a sum projection of the images belonging to the same meristem was computed. On this image, the same pipeline as the one previously described was applied to obtain a 2D parabolic mask outlining the SAM. Then, a second 2D parabolic mask with increased curvature was created based on this and all intensity values of the pixels outside the mask were set to 0. This mask was then divided into consecutive 10-μm sections. This allowed fluorescence intensity concentration profiles to be obtained along the SAM longitudinal axis. Concentration was defined as the ratio of total intensity (sum of pixel intensity) to the total area (sum of the pixel area).
Protein expression and purification for in vitro analysis
Codons of the coding sequences of FD and GRF7 from A. thaliana were optimized to E. coli and cloned into pMAL-c5X-His (NEB) or a modified pMAL-c5X vector. Transformants carrying the recombinant plasmids were grown in LD medium supplemented with appropriate antibiotic to OD600 = 0.6 before induction by 0.6 mM IPTG for 16–20 h at 12 °C. The E. coli cells were collected by centrifugation, resuspended in wash buffer (25 mM Bis-Tris pH 8.0, 160 mM NaCl and 15 mM imidazole) and sonicated to prepare cell lysates. The proteins were purified using Ni-NTA beads (QIAGEN), the bound proteins were washed 5 times with wash buffer and eluted using elution buffer (25 mM Bis-Tris pH 8.0, 160 mM NaCl and 250 mM imidazole). The eluted proteins were further purified by size-exclusion chromatography (HiLoad 16/600 Superdex 200 pg, GE Healthcare) in buffer containing 25 mM Tris-HCl pH 8.0, 160 mM NaCl and 2% (v/v) glycerol.
Structural modelling
The structures of GRF7, full-length FD and truncated and mutant FD were predicted using AlphaFold70 and AlphaFold271. The modelled structure of the FDc–GRF7 and GRF7–FT complex was predicted by ColabFold39. The modelled structure of the FDc–DNA complex was based on a bZIP (PAP1)–DNA complex (Protein Data Bank (PDB): 1GD2)72. The modelled structure of the FAC–DNA complex was based on the modelled FDc–DNA, FDc–GRF7 and GRF7–FT complexes.
SEC–MALS
Purified recombinant proteins were quantified by NanoDrop using the protein-specific extinction coefficient and diluted to the desired concentration (as mentioned in the figures). SEC–MALS was performed in buffer (25 mM Tris-HCl pH 8.0, 160 mM NaCl and 2% (v/v) glycerol) on a 10/600 Superdex 200 pg (home packed) column using an AKTÄ pure 25 M chromatography system coupled to a miniDAWN multi-angle light scattering detector (Wyatt Technology) as well as a refractive index detector (Shodex RI-501). Five-hundred microlitres of sample was used per run at a flow rate of 0.5 ml min−1. BSA was used as a standard for calibration. Baseline correction, selection of peaks and calculation of molecular masses was performed with the ASTRA 8.2 software package.
Gel-shift assay (EMSA)
SEP3 or LFY DNA probe (28 bp) covering one G-box binding site was synthesized by annealing single-stranded 5′-Cy5-labeled oligo in annealing buffer (10 mM Tris pH 8.0, 50 mM NaCl, and 1 mM EDTA pH 8.0). Binding reactions with different proteins with different combinations were indicated in the figures and were carried out in buffer containing 10 mM Tris, 50 ng μL Poly (dI-dC), 50 mM KCl, 10 mM KCl, 1 mM DTT, 5% (v/v) glycerol and 0.1% NP-40. Binding reaction tubes were kept on ice for 20 min and were then loaded onto 6% DNA Retardation Gels (Thermo Fisher) and run in 0.5× Tris/Borate/EDTA buffer at room temperature for 60 min at 100 V. Binding signals were visualized using a ChemiDoc MP Imaging System (Bio-Rad). The primers used for DNA probes are listed in Supplementary Table 4.
Chromatin immunoprecipitation
ChIP methods were described previously with minor modifications60. For ChIP–qPCR of 3HA3Flag-FD, 9 g of above-ground tissue from 10-day-old LD-grown seedlings was collected at ZT 7, and cross-linked for 10 min by vacuum filtration in PBS solution containing 1% formaldehyde. For chromatin immunoprecipitation, 50 μl Dynabeads Protein G beads (Thermo Fisher) coated with 20 μl HA antibody (Abcam, ab9110) was incubated for 4 h with 3 ml of the diluted chromatin solution (1% Triton X-100, 1 mM EDTA, 0.08% SDS, 15 mM Tris-HCl, pH 8.0, and 150 mM NaCl). After washing 3 times with wash buffer (1% NP-40, 1 mM EDTA, 0.1% SDS, 0.1% DOC (sodium deoxycholate, Sigma-Aldrich), 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl), the immune complex was eluted from the beads in 400 μl elution buffer (1% SDS and 0.1 M NaHCO3). Next, samples were reverse cross-linked with 5 μl Proteinase K and 20 μl 5 M NaCl at 65 °C overnight and DNA was purified by a MinElute PCR Purification Kit (QIAGEN). Amounts of input and IP DNA were quantified by fluorometry (Promega, Quantus) and the size of the fragments was analysed by ultra-sensitive capillary electrophoresis (Agilent FEMTOpulse), and the resulting DNA was used for ChIP–qPCR. The primers used for ChIP–qPCR are listed in Supplementary Table 4.
Semi in vivo co-immunoprecipitation and ChIP–qPCR
To perform the semi in vivo co-immunoprecipitation assays for FD and FT, semi-pure nuclei were extracted from 3 g of tissue from 12-day-old FD::3HA-mCherry-FD; fd-3 seedlings grown in long-day conditions as described above. Nuclear proteins were extracted in the sonication buffer (50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 0.5% Triton X-100, 100 mM NaCl, 5 mM MgCl2, 60 mM KCl, 2% (v/v) glycerol, 1 mM PMSF, 50 μM MG132, 1 mM DTT, 1× PIC, 1× PIC2, 1× PIC3). A total of 180 µg E. coli-purified wild-type FT–SII (Strep tag II) or mutated FT–SII protein was added to 800 µl FD nuclear protein, and incubated with rotation for interaction at 4 °C for 2 h.
For co-immunoprecipitation, 30 µl anti-Strep agarose beads (Strep-Tactin XT 4Flow resin, IBA Lifesciences, 2-5010) was pre-equilibrated with the sonication buffer before being added to the protein mixtures. The immunoprecipitation against FT-SII was performed with rotation at 4 °C for 40 min. The beads were washed 4 times in the wash buffer (50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 0.5% Triton X-100, 100 mM NaCl, 5 mM MgCl2, 60 mM KCl, 2% (v/v) glycerol) and then resuspended in 30 µl wash buffer and 10 µl 4× Laemmli loading buffer, followed by boiling for 10 min at 95 °C for immunoblotting analysis using anti-HA–HRP (Roche, 12013819001, 1:1,000 dilution), anti-StrepMAB-Classic–HRP (IBA Lifesciences, 2-1509-001, 1:4,000 dilution) and anti-H3–HRP (Abcam, ab1791, 1:4,000 dilution).
For semi in vivo ChIP–qPCR analysis for purified FT-SII protein, chromatin from 12-day-old FD::3HA-mCherry-FD; fd-3 seedlings grown in long-day conditions was isolated in the above sonication buffer with the addition of 0.24% (w/v) SDS. After interaction of purified FT proteins with the chromatin extractions from in vivo, 0.1 mM disuccinimidyl glutarate followed by 0.1% (v/v) formaldehyde were added to the reaction buffer for 30 min to cross-link FT with interacting proteins and chromatin. ChIP–qPCR was performed as described in ‘Chromatin immunoprecipitation’. Primers are listed in Supplementary Table 4.
Phylogenetic analysis
Nucleotide and protein sequences of bZIP and 14-3-3 gene families were blasted and obtained using ‘makeblastdb’ module in DIAMOND v.2.16.16073. The sequences of both gene families were used for database searching by BLASTP. The initially identified candidate protein sequences in green plants were cut off with an e-value < 10−5. Then, all candidates of bZIP or 14-3-3 families were verified that contained at least one bZIP domain or that were annotated as 14-3-3 proteins by the Conserved Domains Database74 (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). MAFFT75 v.7.490 with auto parameters was used for protein sequence alignment. A maximum likelihood algorithm implemented in IQ-TREE v.1.5.576 with the Jones–Taylor–Thornton model of evolution under GAMMA rate distribution with bootstrapping criterion (up to a maximum of 1,000 bootstraps) was used for phylogenetic analysis. The obtained trees were visualized using the iTOL77 (v.6.7.6; http://itol.embl.de/) phylogeny visualization program.
Ethics and inclusion statement
All data were transparently shared and discussed with all authors during preparation of the manuscript. Authorship criteria were carefully considered, and all contributors were included and their contributions transparently discussed. All materials are available from the authors.
Materials availability
All mutants, transgenic plants, and all plasmid constructions using FD, 14-3-3 and FT genes are available from G.C.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.