Mouse studies
Wild-type C57BL/6 male mice (6–8 weeks of age) were purchased from BIKAI. Nlrx1−/− mice were purchased from Cyagen. NSG mice were purchased from Shanghai Model Organisms Center. All mice were housed in the specific-pathogen-free animal facility of Fudan University with the following environmental parameters: temperature maintained at 21–25 °C, relative humidity at 45–65% and a 12 h–12 h light–dark cycle.
All mice were randomly separated into each experiment group. Mice were fasted from 10:00 for 24 h with free access to water without food. PBS or HC (59847, Sigma-Aldrich; 100 mg per kg) was intraperitoneally injected into mice at 10:00 for 4 h. For acetate administration, mice were fasted for 24 h and PBS or sodium acetate (S5636, Sigma-Aldrich; 1 g per kg) was intraperitoneally injected 10 h and 1 h before mice were euthanized. For serum collection, mice were fasted overnight for 16 h with free access to water. For food reintroduction, mice were fasted for 24 h and then re-fed for another 24 h. Mice were euthanized and the indicated tissues were collected for subsequent analysis.
For the KPC model used in the MRTX1133 therapy experiment, 1 × 106 KPC cells were subcutaneously injected into 6- to 8-week-old NSG mice. The vernier calliper measurements begun when the tumours reached around 200 mm3. Tumour volume measurements were recorded three times per week using the formula 0.5 × length × width2. A blinded study design was used in the mouse tumour experiment to prevent bias during data collection and assessment, thus mice were randomized into control and treatment groups, and treated by intraperitoneal injection with vehicle (10% DMSO + 90% (20% SBE-β-CD in saline) or MRTX1133 in vehicle (30 mg per kg, twice a day) when the tumour volume reached around 300 mm3. Tumours were collected after 6 days of treatment. All of the animal experiment procedures, including the maximal tumour volume, were approved by ethics committee of Department of Laboratory Animals, Fudan University.
AAV production and infection in vivo
Plasmids for the AAV2/9 system, including pAAV RC2/9 plasmids, pAAV helper plasmids and transgene plasmids with the CMV or U6 promoter were used for global expression or the knockdown of genes in vivo respectively as previously described44. The plasmids were mixed with PEI solution and transfected into HEK293T cells. Then, 60–72 h after transfection, the cells and medium were collected by centrifugation (3,500 rpm, 4 °C, 5 min). 5× polyethylene glycol (40% PEG 8000, 2.5 M NaCl) was added to the supernatant and incubated at 4 °C overnight followed by centrifugation (3,000 rpm, 4 °C, 5 min) to collect the virus pellet. Meanwhile, the cell pellet was resuspended with lysis buffer (150 mM NaCl, 20 mM Tris-Cl, pH 8.0) and lysed by three freeze–thaw cycles between liquid N2 and a 37 °C water bath followed by centrifugation (5,500 rpm, 4 °C, 10 min) to obtain the supernatant. Then the supernatant was mixed with the virus pellet. The mixture was purified by Optiprep (D1556-250mL, Sigma-Aldrich) gradients (17%, 25%, 40% and 60%) centrifugation (40,000 rpm, 16 °C, 2 h). The viral fraction was collected from the 40% gradient, then washed three times with PBS using 100 kDa columns (3,500 rpm, 4 °C, 30 min).
AAVs were administered to C57BL/6J mice through gastrocnemius injection (5 × 1010 copies, 25 μl per mouse, three sites) or tail injection (1 × 1011 copies, 150 μl per mouse). All experiments were performed 3–4 weeks after AAV injection. The efficiency of Nlrx1 knockdown or overexpression mediated by AAV delivery was validated by immunoblotting.
Plasmids, reagents and antibodies
Plasmids
WT NLRX1 (HA tag), the NACHT domain (amino acids 160–483, HA tag), LRR domain (amino acids 669–975, Flag tag), ΔLRR (amino acids 1–668, HA tag), 4A (four sites, Glu729, Lys754, Gln758 and Arg958, were mutated to Ala, HA tag) of NLRX1, NLRX1-GFP11 (the C terminus of NLRX1 without stop codon was fused to the linker GGSGGGS and the GFP11 tag RDHMVLHEYVNAAGIT), NLRX1-GFP11-IRES-RFP (the C terminus of NLRX1-GFP11 fused to IRES and RFP), HSP60-GFP11 (the C terminus of HSP60 without stop codon was fused to the linker and the GFP11 tag), GFP11-TOM20 (the GFP11 and the linker fused to the N terminus of TOM20) and HA-NLRP3 were constructed into pcDNA3.1 vector; WT NLRX1-HA, ΔLIR-NLRX1 (amino acid deletion 461–466, HA tag), NLRX1(ΔN-ter) (amino acids 156–975, HA tag), NLRX1(Cyto) (amino acids 87–975, Flag tag), NLRX1(ER) (the N terminus of NLRX1(Cyto) fused to the amino acids 81–250 of FAM134B, Flag tag) were generated as previously described6; cytoGFP(1–10), matrixGFP(1–10) (the N terminus of GFP1–10 was fused to the MTS of COX8, residues 1–36) and GFP-Parkin were generated into the pLVX-hygro vector, and GFP-LC3B was generated into pQCXIH. Constructs encoding mt-Keima and MTS-eGFP were generated into the pLVX or pLVX-Tet-On vector.
Guide RNAs targeting human or mouse NLRX1 were designed online (http://www.e-crisp.org/E-CRISP/) and inserted into the pLentiCRISPR v2 vector.
For NLRX1–HA knock-in cell generation, the guide RNA (5′-TCTGGAAGCTGAGACACTGG-3′) was cloned into the pX458 plasmid. The homology arm of NLRX1-800-stop codon-+800 cloned into pcDNA 3.1 with a mutation at the PAM site from CGG to CCG and HA tag (TACCCCTACGACGTCCCCGACTACGCC) sequence was inserted before the stop codon.
Metabolites
AcCoA sodium salt (AcCoA) (A2056), CoASH (C4780), malonyl-CoA (M4263) and sodium acetate (acetate) (S5636) were obtained from Sigma-Aldrich. Succinly-CoA (HY-137808) was obtained from MCE. Biotin was conjugated to the amino groups (-NH2) of AcCoA by EZ-Link sulfo-NHS-LC-biotin (A39257, Thermo Fisher Scientific) according to the manufacturer’s instructions.
Antibodies
Anti-TIM23 (mouse, 611223, BD Biosciences, 1:5,000), anti-MT-CO2 (rabbit, ab79393, Abcam, 1:3,000), anti-CYTB (rabbit, 55090-1-AP, Proteintech, 1:5,000), anti-HSP60 (goat, sc-13115, Santa Cruz Biotechnologies, 1:1,000), anti-LC3 (rabbit, 3868S, CST, 1:1,000), anti-HA (mouse, 901513, BioLegend, 1:1,000), anti-Flag (mouse, F3165, Sigma-Aldrich, 1:3,000), anti-NLRX1 (rabbit, 17215-1-AP, Proteintech, 1:1,000), anti-PINK1 (rabbit, BC100-494SS, Novus Biologicals, 1:1,000), anti-ACLY (rabbit, 15421-1-AP, Proteintech, 1:1,000), anti-ACSS2 (rabbit, 16087-1-AP, Proteintech, 1:1,000), anti-FASN (rabbit, 10624-2-AP, Proteintech, 1:3,000), anti-ACC1 (rabbit, 21923-1-AP, Proteintech, 1:2,000), anti-IDH1 (rabbit, 12332-1-AP, Proteintech, 1:3,000), anti-p62 (rabbit, 18420-1-AP, Proteintech, 1:3,000), anti-acetylated-lysine (rabbit, 9441, CST, 1:1,000), anti-phospho-AMPKα (rabbit, 40H9, CST, 1:1,000), anti-AMPKα (rabbit, 10929-2-AP, Proteintech, 1:3,000), anti-ULK1 (rabbit, 8054, CST, 1:1,000), anti-phosphorylated ULK1 (Ser757) (rabbit, 6888, CST, 1:1,000), anti-phosphorylated ULK1 (Ser555) (D1H4) (rabbit, 5869, CST, 1:1,000), anti-S6 kinase (S6K) (rabbit, 9202, CST, 1:1,000), anti-phospho-S6K (Thr389) (mouse, 9206, CST, 1:1,000), anti-cytochrome c (rabbit, 556432, BD Biosciences, 1:1,000), anti-Parkin (rabbit, Proteintech, 66674-1-Ig, 1:500), anti-CLIMP63 (mouse, ENZ-ABS-669-0100, ENZO, 1:500), anti-FIP200 (rabbit, 17250-1-AP, Proteintech, 1:3,000), anti-ATG7 (rabbit, 10088-2-AP, Proteintech, 1:3,000), anti-tubulin (rabbit, 11224-1-AP, Proteintech, 1:5,000), anti-actin (mouse, 66009-1-Ig, Proteintech, 1:5,000) were used in immunoblotting. Anti-LC3 (rabbit, PM036, MBL, 1:100), anti-HA (mouse, 901513, BioLegend, 1:1,000), anti-TOM20 (mouse, 612278, BD Biosciences, 1:1,000) and anti-HSP60 (goat, sc-13115, Santa Cruz Biotechnologies, 1:1,000) were used in immunofluorescence. The fluorescent secondary antibodies goat anti-mouse Alexa Fluor 594 (A11032, Invitrogen, 1:1,000), donkey anti-rabbit Alexa Fluor 594 (A21207, Invitrogen, 1:1,000), donkey anti-mouse Alexa Fluor 488 (A21202, Invitrogen, 1:1,000) and donkey anti-goat Alexa Fluor 647 (A21447, Invitrogen, 1:1,000) were used in immunofluorescence.
Inhibitors
HC (59847), BTC (51520) and CCCP (C2759) were from Sigma-Aldrich. SB (HY-16450), actinomycin D (HY17559), MRTX1133 (HY-134813), RMC-6236 (HY-148439), Torin-1 (HY-13003) and Mdivi-1 (HY-15886) were from MCE. Bafilomycin A1 (S1413) was from Selleck. Oligomycin (9996) was from CST.
Cell culture and cell line generation
HEK293T, HeLa, A549, MCF7 and U-2 OS cells were purchased from ATCC and AsPC-1 cells were purchased from National Collection of Authenticated Cell Cultures (NCACC), Shanghai. Sf9 cells were purchased from Invitrogen. KPC (KrasG12D/+Trp53R172H/+) cells were obtained from Z.-G. Zhang. HEK293T, HeLa, A549, MCF7, U-2 OS, KPC cells and AsPC-1 cells were cultured in DMEM (Invitrogen) or RPMI-1640 (Invitrogen) supplemented with 10% FBS (BI) and 1% penicillin–streptomycin (HyClone). Sf9 cells were cultured in SF900 II SFM (Gibco). The SM is the DMEM formula with 5 mM glucose, 2 mM glutamine, 1 mM pyruvate and supplemented with 10% dialysed serum (BI) and 1% penicillin–streptomycin (HyClone). All cell lines were grown at 37 °C and 5% CO2 and were tested to be mycoplasma free using the mycoplasma detection kit (40612ES25, YEASEN). A notable exception was the Sf9 cell line, which was maintained under distinct conditions: incubation at 28 °C with shaking on a horizontal shaker at a rotational speed of 100 rpm.
NLRX1-knockout cells were generated using the CRISPR–Cas9 system. pLentiCRISPR v2 vectors carrying sgRNA were mixed in Opti-MEM and transfected into cells with polycation polyethylenimine (PEI) (Sigma-Aldrich) and selected by puromycin for 3 days to get NLRX1-deficient cells. Single cells were seeded into 96-well plates and validated by sequencing and immunoblotting to get NLRX1-knockout cells. To generate NLRX1–HA-tag knock-in cells (HeLa-NLRX1(HA-KI)), the plasmid pX458 together with donor DNA (amplification of plasmid pcDNA3.1 containing the homology arm of NLRX1) with a ratio of 1:1 in Opti-MEM were transfected with Lipo3000 (Invitrogen) into HeLa cells. After 48 h transfection, GFP-positive cells were sorted and seeded into the 96-well plate (single clone per well) by flow cytometry. The knock-in cells were validated by sequencing and immunoblotting.
To generate cells with the inducible expression of mt-Keima, HeLa cells were infected with viruses expressing pLVX-Tet3G-rtTA and selected with G418 (800 μg ml−1) for 1 week to get HeLa-rtTA cells. HeLa-rtTA cells were then infected with viruses expressing mt-Keima, followed by blasticidin (10 μg ml−1) selection for an additional 1 week to generate HeLa-Tet-On-mt-Keima cells. The expression of mt-Keima was induced by doxycycline (1 μg ml−1) for 6 h.
To generate stable cell lines, cells were infected with indicated viruses together with 10 μg ml−1 polybrene. After 48 h, cells were selected with 2 μg ml−1 puromycin, 50 μg ml−1 hygromycin, 10 μg ml−1 blasticidin or 800 μg ml−1 G418 for 1–2 weeks. The overexpression or knockdown efficiency was verified by immunoblotting.
For transient gene overexpression, cells were transfected with indicated plasmids using Lipo3000 (for HeLa cells) or PEI (for HEK293T cells). Gene expression was validated by immunoblotting or immunofluorescence 24–48 h after transfection.
Virus packing
Lentiviral or retroviral vectors carrying the indicated genes, together with packaging plasmids psPAX2 and pMD2.G or VSVG and GAG were transfected into HEK293T cells. After 48–72 h, the supernatants were collected, filtered with a 0.45-μm filter and concentrated with PEG8000.
Gene knockdown by siRNA
siRNAs were transfected by Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions and, after 48 h, the transfected cells were treated as indicated and collected for subsequent analysis.
Mitochondria isolation
For mitochondria isolation, cells were washed with cold PBS twice and collected with 1 ml cold mitochondrial lysis buffer as previously described45. The buffer containing 210 mM mannitol, 70 mM sucrose, 5 mM Tris–HCl (pH 7.5) and 1 mM EDTA (pH 8.0), was adjusted to pH 7.5 with protease inhibitors. Then the cell suspension was transferred to the Dounce Tissue Grinder (P1110, T2690, Sigma-Aldrich) and lysed by 26 strokes. The homogenate was centrifuged at 1,300g for 10 min at 4 °C and the supernatant was collected in new tubes followed by centrifugation (10,000g, 4 °C, 20 min) to generate the supernatant as the cytosolic fraction and the cell pellet as the mitochondrial fraction.
Immunoblotting, immunoprecipitation and GST-pull-down assay
For immunoblotting, cells were lysed in 1× SDS buffer, boiled at 95 °C for 10 min and analysed by SDS–PAGE. For LC3 analysis, cells were lysed by buffer F (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 0.5% NP-40 and protease inhibitors) and centrifuged for 15 min at 4 °C. The supernatants were collected and boiled with 3× SDS and analysed by SDS–PAGE. For biotin–AcCoA pull-down assays, cells were collected and mitochondria were purified as described above. Mitochondria pellets were lysed by buffer C (50 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 2 mM EDTA and protease inhibitors) and centrifuged for 15 min at 4 °C; the supernatants were then collected for the biotin–AcCoA-binding assay. Streptavidin beads (3419, CST) were incubated with biotin or biotin-labelled AcCoA in PBS for 1 h at room temperature, the beads were washed once with PBS and then incubated with cell lysates overnight at 4 °C with rotation. On the second day, the beads were washed four times with buffer C, boiled with 1× SDS and analysed by SDS–PAGE. For GST–LC3 pull-down assays, GST beads (AGM90049, AOGMA) were incubated with recombinant GST–LC3 for 4 h at 4 °C, then washed with buffer C and incubated with cell lysates at 4 °C for 4 h with rotation. The beads were washed four times with buffer C, boiled with 1× SDS and analysed by SDS–PAGE. For LRR-domain binding with the NACHT domain, cells were lysed with lysis buffer (0.5% Triton X-100, 20 mM HEPES pH 7.6, 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA with protease inhibitors). AcCoA was co-added with Flag beads (A2220, Sigma-Aldrich) into the lysates overnight at 4 °C with rotation. The beads were washed and protein samples were processed as described above.
For AMPK activation and mTOR inhibition analysis, HeLa, A549 and MCF7 cells were treated with SM (DMEM containing 5 mM glucose, 2 mM glutamine and 1 mM pyruvate sodium with 10% dialysed serum and 1% penicillin–streptomycin) for 16 h. Oligomycin (5 μM, 5 min) or Torin-1 (100 nM, 16 h) was used as a positive control. Cells were washed with precooled PBS twice and lysed with precooled lysis buffer containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM pyrophosphate, 50 mM NaF, 5 mM β-glycerol phosphate, 50 nM calyculin A, 1 mM Na3VO4 and protease inhibitors. The lysates were centrifuged at 17,000g for 10 min at 4 °C, and the supernatant was boiled with 3× SDS and analysed by SDS–PAGE46.
For all immunoblotting analyses, proteins were separated by 10–12% SDS–PAGE, transferred to PVDF membranes (Amersham), blocked by 5% non-fat milk in 0.1% Tween-20/PBS buffer for 1 h at room temperature, and immunoblotted by antibodies according to molecular mass. All uncropped raw immunoblotting data are provided in Supplementary Fig. 1.
Purification of recombinant NLRX1 proteins
Human NLRX1 without the N-terminal MTS (amino acids 87–975) was cloned into the pFastBac His vector with an additional N-terminal MBP tag. The vector was then transfected into DH10Bac to get recombinant bacmids, which were further transfected into SF9 insect cells to get amplified baculovirus. SF9 cells were infected with amplified baculovirus for 3 days and cells were collected and lysed in HEPES buffer (20 mM HEPES pH 7.5, 150 mM NaCl) with protease inhibitors and 0.5 mM Tris (2-carboxyethyl) phosphine (TCEP) with sonication. After centrifuging with 10,000 rpm for 1 h, supernatants containing recombinant NLRX1 were purified with Ni-NTA (QIAGEN) and gel-filtration chromatography on the Superdex 200 column (GE Healthcare). The purified recombinant MBP–NLRX1–His was confirmed by immunoblotting using NLRX1 antibody and used for biotin–AcCoA pull-down assay.
NLRX1 LRR domain (629–975) or LRR(4A) were cloned into the pMAL-c5X vector with an N-terminal expressed MBP tag. Constructs were transfected into Escherichia coli BL21 (DE3) cells, which were incubated in LB medium (50 μg ml−1 ampicillin) for 6 h at 37 °C with shaking. Protein expression was induced with 0.2 mM isopropyl-β-D-thiogalactopyranoside overnight at 18 °C. Cells were collected and resuspended in HEPES buffer with protease inhibitors and 0.5 mM TCEP. The proteins were further purified by gel-filtration chromatography on the Superdex 200 column (GE Healthcare Life Sciences) equilibrated with the HEPES buffer with 0.5 mM TCEP. Dextrin beads (SA077025, Smart-Lifesciences) were used to purify recombinant proteins, washed with HEPES buffer and eluted with 5 mM maltose.
[3H]AcCoA binding assay
Recombinant MBP–NLRX1-LRR proteins (50 μg) and equal amounts of MBP or MBP–LRR(4A) proteins were incubated with dextrin beads for 2.5 h at 4 °C with rotation. Beads were washed twice with lysis buffer 2.0 (HEPES buffer with 2 mM MgCl2, 0.5 mM TCEP and 0.05% Tween-20). The beads were incubated with 2 μM [3H]AcCoA (NET290250UC, Perkin Elmer) and the indicated concentrations of cold AcCoA for 1 h at room temperature. The tubes were flicked every 10 min. The beads were then washed four times with lysis buffer 2.0 and quantified using the TriCarb scintillation counter (PerkinElmer). The binding affinity Kd was calculated as previously described47.
Protein oligomerization analysis
Protein oligomerization analysis was conducted as previously described6,48. Cells were washed twice with PBS and centrifuged at 3,000 rpm for 5 min at 4 °C. After resuspending in PBS, cell pellets were pipetted 24 times with a 22-gauge needle and centrifuged at 13,000 rpm for 1 h at 4 °C followed by gentle sonication in PBS. The samples were divided into two parts—one part was cross-linked with 1 mM glutaraldehyde for 10 min at 16 °C and the other was not cross-linked as inputs. The samples were boiled at 95 °C for 10 min and analysed by immunoblotting after SDS–agarose or SDS–PAGE electrophoresis.
Molecular modelling for AcCoA binding to NLRX1
The NLRX1 structure was from the Protein Data Bank31 (PDB: 3UN9). The simulation of AcCoA or CoASH docking to LRR of NLRX1 was performed by Schrödinger Computational Suite, Maestro v.11.5.011, MMshare v.4.1.011, release 2018-1, platform Windows-x64. All structure figures were prepared in Pymol (http://www.pymol.org).
Flow cytometry
Cells were treated as indicated and washed with PBS, collected in DMEM or RPMI1640 and centrifuged at 800g for 5 min. Cells were stained with 100 nM TMRM (T668, Invitrogen), 5 μM MitoSOX (M36008, Invitrogen) or 10 μM CM-H2DCFDA (HY-D0940, MCE) in DMEM or RPMI1640 for 30 min at 37 °C and 5% CO2. After the incubation, cells were washed twice with PBS and resuspended in DMEM or RPMI1640 followed by flow cytometry.
The split GFP system used for monitoring mitochondrial outer membrane or matrix protein localization has been previously described27,49,50. In brief, HeLa cells stably expressing cytoGFP(1–10) or matrixGFP(1–10) were transfected with construct encoding NLRX1(G11)-IRES-RFP. After 36–48 h, cells were treated as indicated and flow cytometry was performed by Beckman coulter CytoFLEX S instrument. NLRX1 localization on the mitochondrial outer membrane or matrix was calculated on the basis of the GFP+RFP+/RFP+ ratio. For flow cytometry, 1 × 104 to 2 × 104 cells were collected using the Beckman Courtier instrument, and the data were analysed by FlowJo v.10.8.1 software or CytExpert 2.5.
ATP measurement and cell death assay
For intracellular ATP production measurement, the ATP Assay Kit (S0026, Beyotime) was used according to the manufacturer’s instructions. In brief, cells were washed twice with PBS and lysed with lysis solution for 20 min on ice followed by centrifugation (12,000g, 5 min, 4 °C). ATP assay working solution and the supernatants were co-added into a black 96-well plate. The luminescence was measured using a microplate reader (BioTek).
For the cell death assay, LDH was detected using the CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (G1782, Promega) according to the manufacturer’s instructions. In brief, cells were treated as indicated and the supernatants were transferred to a fresh 96-well plate. An equal volume of LDH detection working solution was added to each well plate and incubated at 37 °C for 30 min. The LDH positive control in the kit was used as the positive control. Finally, the absorbance signal was measured at 490 nm using a microplate reader (BioTek).
Measurement of NADP(H)
For the measurement of NADP(H), the NADP+/NADPH Assay Kit (Beyotime, S0180) was used according to the manufacturer’s instructions. In brief, cells were washed twice with PBS, resuspended with extraction buffer and then centrifuged (12,000g, 5 min, 4 °C). The supernatant was divided into two equal parts. One part was used for total NADPH measurement. The other part was incubated for 30 min at 60 °C to decompose NADP+. The G6PDH assay working solution was then added to the supernatants in the 96-well plate and the mixture was incubated for 10 min. Finally, the absorbance signal was measured at 450 nm with a microplate reader (BioTek).
2D cell proliferation
For 2D cell proliferation assay, cells were seeded into the black 96-well plate and treated with serial dilutions of MRTX1133 or RMC-6236, Mdivi-1 (20 μM) or DMSO. Then, 50 μl of CellTiter-Glo reagent was added to 50 μl of medium-containing cells and the contents were mixed for 2 min. The plate was next incubated at room temperature for 10 min. The luminescence was measured using a microplate reader (BioTek). Luminescence signal was normalized to DMSO treated cells (percentage DMSO = (lumtreated/mean(lumDMSO)) × 100). A log[inhibitor] versus response-variable slope (four parameters) model was used to calculate the IC50.
Metabolite extraction and GC–MS
For intracellular metabolite measurements in Extended Data Fig. 1n, cells were cultured in 10 cm dishes and treated with SM for 16 h. When cell confluency was about 80–100%, the medium was removed, cells were washed with cold PBS twice, collected in extraction buffer (acetonitrile:isopropanol:water, 3:3:2, v/v/v). The resuspended cells were placed in liquid N2 for 5 min and thawed on ice for 5 min and the freeze–thaw cycle was repeated four times and the samples were then centrifuged (12,000 rpm, 4 °C, 10 min) to collect the supernatants. For serum valine, leucine and isoleucine measurements in Extended Data Fig. 1c, blood was collected from the eyes and clotted at room temperature for 1 h and then centrifuged (3,000 rpm, 10 min). Next, 20 μl serum was diluted with 80 μl precooled methanol and vortexed, then centrifuged at 12,000g for 15 min to remove proteins. The cell or serum supernatants were then evaporated by freeze-vacuum and analysed by gas chromatography coupled with MS (GC–MS). The pellets were resuspended in 75 μl acetonitrile at 60 °C for 15 min, then oximated by MTBSTFA (N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide) in 50 μl pyridine and further incubated in 60 °C for 1 h. The samples were centrifuged and the supernatants were transferred to glass vials. Then, 1 µl of each sample was injected and analysed on the Agilent 7890B-5977B GC–MS system with DB-5MS (0.25 mm internal diameter, 0.25 μm film, with 30 m empty column, Agilent J&W). Metabolite m/z ratios were compared with those of previous studies51. Each metabolite was quantified by the retention time and peak area by MassHunter Workstation (Agilent). The final intracellular metabolites and amino acid level was normalized to actin level of immunoblotting or volume of serum.
Metabolite extraction and LC–MS
Cytosolic and mitochondrial AcCoA levels were determined as previously described45. In brief, approximately 1 × 107 to 2 × 107 cells were lysed with 1 ml cold mitochondrial lysis buffer and the cytosolic and mitochondria fractions were then isolated as described above. The cytosolic fraction was quenched by 50% (w/v) trichloroacetic acid in water (the final concentration of trichloroacetic acid is 10%) and the mitochondria fraction was resuspended in 1 ml 10% (w/v) trichloroacetic acid. The mitochondria fraction was placed into liquid N2 for 5 min and thawed on ice for 5 min and the freeze–thaw cycle was repeated four times followed by centrifugation (17,000g, 4 °C, 10 min). The mitochondria supernatants and cytosolic fractions were then purified using Oasis HLB 1cc (30 mg) SPE columns (Waters). Columns were washed with methanol, equilibrated with water, loaded with the cytosolic and mitochondrial fractions, washed with water and eluted with elution buffer (25 mM ammonium acetate in methanol). The elutions were evaporated by freeze-vacuum, resuspended in 20 μl 20% acetonitrile in water and analysed by LC–MS. For in vivo AcCoA measurement, tissues were obtained from C57BL/6J mice, and 50–100 mg tissue was collected in 1 ml precooled 10% trichloroacetic acid. The samples were then homogenized and lysed on a rotating shaker (30 min, 4 °C) followed by centrifugation (12,000g, 10 min, 4 °C). The supernatant was purified, dried and resuspended as described above, and analysed using LC–MS. Then, 5 µl of each sample was injected and analysed using SHIMADZU LC-30AB LC system coupled to the QTRAP7500 Mass Spectrometer (SCIEX). Hydrophilic interaction chromatography (HILIC) with the BEH column (1.7 µm, 2.1 mm × 100 mm; Waters) was used. Mobile phase A was as follows: ammonia with 10 mM ammonium formate and 0.2% ammonia. Mobile phase B was acetonitrile. The flow rate was 0.2 ml min−1 and the column temperature was set at 40 °C. Linear gradient: 0 min, 80% B; 3 min, 50% B; 10 min, 50% B; 10.1 min, 80% B; 15 min, 80% B. Multiple reaction monitoring (MRM) technology using MS/MS was used for specific detection of AcCoA. The LC–MS system was operated in negative ionization mode. The source parameters included curtain gas (CUR) at 40 psi; collision active dissociation (CAD) gas at 6; ion source gas 1 (GS1) at 40 psi, GS2 at 70 psi; ion spray voltage (IS) at 4,500 V; ion source temperature (TEM) at 450 °C. The specific transition was recorded as follows: AcCoA 808.0945 > 407.9000. The final AcCoA level was normalized to the actin immunoblot level, or the tissue weight or cell number, as described in the figure legends.
For serum amino acid measurement (proline, glutamate acid, serine, asparagine, glutamine, arginine, glycine, alanine, aspartic acid, tyrosine, histidine, lysine, methionine, phenylalanine, threonine and tryptophan, while cysteine was too low to be detected), blood was collected from eyes and clotted at room temperature for 1 h followed by centrifugation (3,000 rpm,10 min). Then, 20 μl serum was diluted with 80 μl precooled methanol and vortexed, then centrifuged at 12,000g for 15 min to remove proteins. The supernatants were evaporated by freeze-vacuum, resuspended in 100 μl 80% methanol in water and analysed by LC–MS. Then, 1 µl of each sample was injected and analysed using UPLC-H Class LC system (Waters) coupled to the 6500 QTRAP Mass Spectrograph (SCIES). The ultimate AQ-C18 column (5 µm, 2.1 mm × 250 mm; Welch) was used at room temperature. Mobile phase A was as follows: water (0.1% formic acid, v/v); and mobile phase B was acetonitrile (0.1% formic acid, v/v). Linear gradient: 0–1 min, 0% B; 1–14 min, 0–90% B; 14–16 min, 90% B; 16–16.1 min, 90–0% B; 16.1–20 min, 0% B. The flow rate was 0.2 ml min−1. MRM technology using MS/MS was used for specific detection of various amino acids. The LC–MS system was operated in positive ionization mode. The source parameters included CUR at 40 psi; CAD at medium; GS1 at 40 psi, GS2 at 40 psi; IS at 5,500 V; and TEM at 500 °C. All LC–MS analysis was performed by the Metabolic Platform at the Fudan University. Data were analysed using Skyline (22.2.0.527) software to calculate the peak area values.
Seahorse analysis
The mitochondrial oxygen consumption rate (OCR) in HeLa cells was measured with the Seahorse XFe96 equipment (Agilent) using the Cell Mitochondrial Stress Test kit (103015-100, Agilent) according to the manufacturer’s instructions. In brief, 0.8 × 104 cells were seeded onto an XFe96 cell culture microplate (Agilent) per well and treated with CCCP (10 μM), SB (100 μM), BTC (5 mM), HC (20 mM) and SM for 16 h. Before analysis, cells were washed twice and equilibrated with XF DMEM in a 37 °C incubator without CO2 for 1 h. Oligomycin (final concentration: 1.5 μM), FCCP (final concentration: 2 μM), and rotenone/antimycin A (final concentration: 0.5 μM) were used in OCR analysis. Data were analysed by Seahorse Wave Desktop Software (Agilent).
CRISPR screening
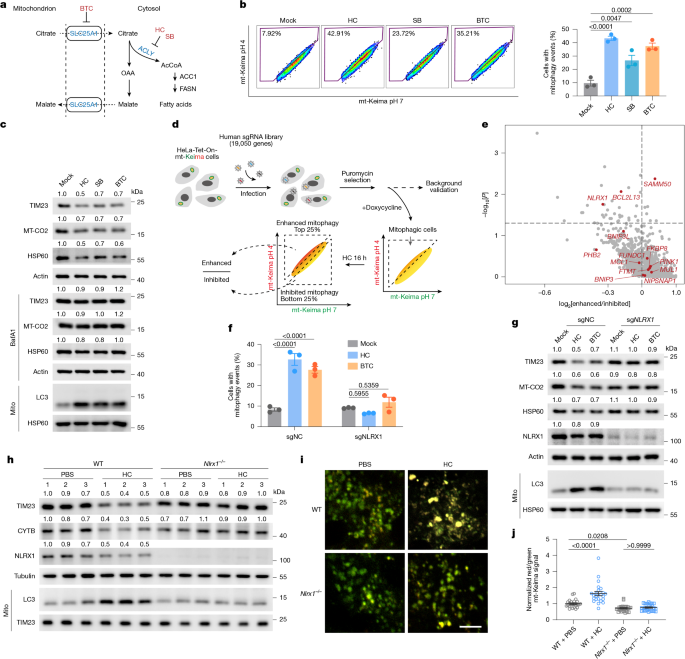
To generate lentivirus for screening, we used the genome-wide GeCKO v2.0 Human library52 in the lentiCRISPR v2 vector (Addgene, 1000000048), which contains six sgRNAs per gene (123,411 sgRNAs targeting 19,050 genes). A total of 1 × 108 HEK293T cells was seeded into six T225 flasks. Each flask was transfected with 20 μg of plasmid library, 10 μg of psPAX2 and 5 μg of pMD2.G using PEI. After 48 h of transfection, the supernatant was collected, centrifuged at 3,000 rpm for 5 min, filtered through a 0.45-μm filter and stored at −80 °C. Virus titres were determined using puromycin selection. The lentiviral library was used to infect HeLa cells stably transfected with doxycycline-inducible mt-Keima reporter at a multiplicity of infection of approximately 0.3 with 8 μg ml−1 polybrene. Then, 48 h after infection, 1 μg ml−1 puromycin was added to the cells and selected for 5 days, followed by an additional 2 days of expansion in puromycin-free medium. During selection, cells were maintained at >500 cells per sgRNA.
To induce mitophagy, cells were first treated with 1 μg ml−1 doxycycline overnight to induce mt-Keima expression, followed by 20 mM HC treatment for 16 h. Treated cells were trypsinized, filtered through a 40-μm cell strainer and resuspended in PBS containing 2% FBS. Cell sorting was performed using a BD FACSAria II instrument with two channels: 405 nm excitation for mt-Keima at pH 7 and 562 nm excitation for mt-Keima at pH 4, with a 610 nm emission bandwidth53. The top 25–30% and bottom 25–30% cells were sorted to represent mitophagy-enhanced and -inhibited cell subsets, respectively. A total of 107 cells was sorted for each group in two biological replicates for subsequent sequencing.
Genomic DNA was extracted from both the mitophagy-enhanced and -inhibited groups. Sequencing libraries were prepared by two rounds of PCR to amplify target DNA fragments, followed by the ligation of index and adapter sequences. The prepared libraries were then subjected to paired-end sequencing (2 × 150 bp) on the Illumina NovaSeq 6000 platform. After sequencing, 20 bp gRNA sequences were extracted and aligned to the GeCKO v2.0 library reference sequence. The alignment results for all library sequences were counted to obtain the number of matched reads. Sequencing depth and coverage were calculated to assess data reliability and accuracy. Two rounds of screening data were analysed using MAGeCK software54 with the default settings, and the results were ranked by negative log2-transformed fold change and P value. Mitochondrial genes were defined using the mitoCarta3.0 database22. Volcano plots for genome-wide and mitochondria-targeted analyses of the CRISPR screening were generated using the R package ggplot2. GO biological processes enriched in the top 100 mitochondrial genes from the screen were analysed using the Metascape database55.
qPCR analysis
To quantify the mtDNA/nDNA ratio, genomic DNA was isolated from cells or tissues using the TIANamp Genomic DNA Kit (DP304-02, TIANGEN) according to the the manufacturer’s instructions, and qPCR was conducted to amplify the mitochondria genome (MT-CYTB, MT-CO1, MT-ATP6 in human; mt-Cytb, mt-Co1, mt-Atp6 in mouse) or nuclear genome (RPL13A in human; Rpl13a in mouse) separately as previously described6. Total RNAs were extracted using the RNA Easy Fast Tissue/Cell Kit (TIANGEN, DP451) and reverse transcribed using the PrimeScript RT Reagent kit (TaKaRa, RR047A) according to the manufacturer’s instructions. The qPCR was performed on the ABI QuantStudio 7 Flex system using the TB Green Premix ExTaq kit (TaKaRa, RR820A). The relative fold changes were calculated using \({2}^{-\Delta \Delta {C}_{{\rm{t}}}}\) method. qPCR primer sequences are provided in Supplement Table 3.
Immunofluorescence and confocal microscopy
For colocalization analysis of exogenous NLRX1 with exogenous LC3, HeLa cells stably expressing NLRX1 with a C-terminal HA tag (HeLa-NLRX1-WT) were cultured on glass coverslips and transfected with the vector expressing GFP–LC3. For colocalization analysis of endogenous NLRX1 with endogenous LC3, HeLa cells with HA tag knock-in (HeLa-NLRX1(HA-KI)) were used. For colocalization analysis of GFP–Parkin with mitochondria, HeLa cells stably expressing GFP–Parkin were generated (HeLa-GFP–Parkin). Cells were treated as indicated in the figure legends and washed with PBS twice, fixed with 4% PFA for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, blocked with 5% BSA in PBS for 1 h at room temperature and incubated with primary antibodies (diluted in 5% BSA) at 4 °C overnight. The next day, cells were washed three times with PBS and incubated with fluorescent secondary antibodies for 2 h at room temperature and then washed three times with PBS. After incubation with DAPI for 5 min and mounted with antifade reagent, samples were observed with the ×60 oil objective of confocal microscopy (Leika SP5 and Olympys FV3000).
For analysis of mitochondrial protein import, HeLa-rtTA cells were transfected with vectors expressing Tet-On-MTS-EGFP. After 6 h of transfection, 0.25 μg ml−1 doxycycline was added to induce MTS-EGFP expression. Then cells were stained with 50 nM Mitotracker Deep Red FM (M22426, Thermo Fisher Scientific) for 20 min and washed with PBS twice, and fresh medium was added for living cell imaging using Olympys confocal microscope (FV3000) with a ×60 oil objective in two channels: 488 nm excitation and 520 nm emission for eGFP and 640 nm excitation and 685 nm emission for Mitotracker Deep Red FM. Three replicates with a total of 100 cells per condition were analysed29,56.
The split GFP system used for monitoring mitochondrial outer membrane or matrix protein localization was described above. In brief, HeLa cells stably expressing cytoGFP(1–10) or matrixGFP(1–10) were transfected with constructs encoding NLRX1-GFP11, HSP60-GFP11 or GFP11-TOM20 vectors. After 24 h, living cells were observed using Olympus confocal microscope (FV3000) with a ×60 oil objective in the channel with 488 nm excitation and 520 nm emission for GFP. Images were processed by deconvolution using OLYMPUS CellSens Dimension Desktop (v.4.1.1) to improve resolution, remove background fluorescence and recover the real distribution.
Mitophagy reporter assay
For measuring in vivo mitophagy, 6–8-week-old mice were given intramuscular injection of AAV-mt-Keima (3 × 1011 copies, 25 μl per mouse, three sites) or intravenously (1 × 1011 copies, 150 μl per mouse). After 4–6 weeks, mice were fasted for 24 h or given intraperitoneal injection of HC for 4 h, and gastrocnemius or liver tissues were collected. The samples were cut into sections with a thickness of 6 μm and observed using Olympus confocal microscope (FV3000) with a ×60 oil objective in two channels: 445 nm excitation for mt-Keima pH 7 and 561 nm excitation for mt-Keima pH4 with a 570–695 nm emission bandwidth. For mitophagy index quantification, total mitochondrial area and mitolysosome area were calculated by green fluorescence area and red-only puncta area gated by a fixed threshold individually using Image J software. Mitophagy level was quantified by the ratio of mitolysosome area/mitochondrial area.
For measuring in vitro mitophagy, cells were treated as indicated, and the flow cytometry was performed by Beckman coulter CytoFLEX S instrument in two channels (BV605 and PE-Texas Red channels) through two lasers (405 nm and 562 nm) and emission at 610 nm. The gating strategy was provided in Supplementary Fig. 2. Data were analysed using CytExpert 2.5. The ratio of the mitophagic percentage was calculated and the quantification was pooled from three independent biological replicates.
Cytochrome c release analysis
HeLa cells were treated as indicated, washed with cold PBS and lysed with digitonin lysis buffer (150 mM NaCl, 50 mM HEPES pH 7.4, 25 μg ml−1 digitonin with protease inhibitors) for 10 min on ice. The lysates were then centrifuged at 2,000g for 10 min at 4 °C. The supernatants were centrifuged at 20,000g at 4 °C twice, and the final supernatant was the cytosolic fraction for the detection of cytosolic cytochrome c57,58.
Tissue mitochondrial isolation
Cytosolic and mitochondrial fraction isolation was determined as previously described20. In brief, for mitochondria isolation from gastrocnemius tissue, mice were killed and muscles were removed, washed with cold PBS supplemented with 10 mM EDTA and minced. The muscles were incubated with PBS supplemented with 10 mM EDTA and 0.05% trypsin for 30 min followed by centrifugation (200g, 10 min, 4 °C). The pellet was resuspended with IBm1 (67 mM sucrose, 50 mM Tris/HCl, 50 mM KCl, 10 mM EDTA and 0.2% BSA, pH adjusted to 7.4) and transferred to the Dounce Tissue Grinder and lysed by 15 strokes followed by centrifugation (700g, 10 min, 4 °C). The supernatant was then centrifuged at 8,000g for 10 min at 4 °C. The supernatant is the cytosolic fraction. Then pellet was resuspended with IBm2 (0.25 M sucrose, 3 mM EGTA/Tris and 10 mM Tris/HCl, pH adjusted to 7.4) followed by centrifugation (8,000g, 10 min, 4 °C) to obtain the mitochondrial pellet.
For mitochondria isolation from liver, mice were killed and livers were collected, washed with cold IBc (10 mM Tris–MOPS, 1 mM EGTA/Tris, 0.2 M sucrose, pH adjusted to 7.4) and minced. Then the liver suspension was transferred to the Dounce Tissue Grinder and lysed by ten strokes, followed by centrifugation (600g, 10 min, 4 °C). The supernatant was then centrifuged at 7,000g for 10 min at 4 °C. The supernatant was the cytosolic fraction. The pellet was then resuspended with IBc followed by centrifugation (7,000g, 10 min, 4 °C) to obtain the mitochondrial pellet. For LC–MS, both the cytosolic and mitochondrial fractions were performed as described above. For immunoblotting analysis, both the cytosolic and mitochondrial fractions were boiled with 3× SDS and analysed by SDS–PAGE.
Electron microscopy
HeLa cells were treated with indicated conditions, transformed into suspension cells using a cell shovel and centrifuged at 2,000 rpm for 10 min. The pellets were fixed in 2.5% glutaraldehyde for 1 h at room temperature and then overnight at 4 °C. The next day, after washing three times with 0.1 M PBS, the pellets were fixed with 1% osmic acid at room temperature for 1 h, washed three times with double-distilled H2O, dehydrated in a graded ethanol series, slowly infiltrated with 100% acetone and 50% acetone (acrylic resin: acetone,1:1, v/v) for 2 h and embedded in acrylic resin at 60 °C for 48 h. The embedded samples were cut into sections with a thickness of 70 nm, and the sections were stained with 2% uranyl acetate at room temperature for 10–20 min and lead citrate stain the sections for 5 min. Lastly, samples were observed by electron microscope and images were captured by FEI Tecnai G2 spirit electron microscope.
Thermal shift assay
HeLa cells were lysed using lysis buffer and centrifuged at 12,000 rpm for 10 min at 4 °C. PBS, AcCoA (500 µM) or CoASH (500 µM) were added to cell lysates, heated to graded temperatures (44.6–65 °C, 3 min) and centrifuged at 12,000 rpm for 10 min at 4 °C. Soluble proteins were extracted and analysed by immunoblotting.
Statistical analysis
All data were analysed using GraphPad Prism (v.8.3.0) or Excel, with n ≥ 3 biological replicates unless otherwise specified. Data are presented as mean ± s.e.m. or mean ± s.d. as indicated. For the CRISPR screen data in Fig. 1e and Extended Data Fig. 5k, n = 2 biological replicates. All statistical tests were two-tailed. Statistical parameters, including scale bars and statistical significance, are shown in the figures and the figure legends. Two-group comparisons were analysed using unpaired t-tests. Multiple comparisons among more than two groups were performed using one-way ANOVA. A two-way ANOVA was used when two categorical variables were analysed. Post hoc analysis was conducted to identify specific group differences following ANOVA. P < 0.05 was considered to be statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.