Cell lines
KP LUAD cell lines were obtained from the laboratory of T. Jacks. KP, Fsp1KO cell lines were generated by transient transfection of PX458 (Addgene #48138) expressing an sgRNA targeting Fsp1. Single GFP-positive clones were selected and FSP1 loss was validated by western blot. The 16645 cell line was developed from KrasG12D Stk11−/− GEMM as previously described52. KPC7 cells were obtained from the laboratory of D. Simeone. All cell lines were maintained in DMEM or RPMI 160 (Corning) supplemented with 10% FBS (Sigma Aldrich) and gentamicin (Invitrogen) and were tested for mycoplasma regularly (PlasmoTest, InvivoGen). All mouse cell lines were authenticated by PCR genotyping. All human cell lines used were purchased from ATCC and were authenticated by short-tandem-repeat profiling. Genetic manipulation of cell lines was performed via lentiviral transduction of plasmids, detailed in next section, followed by either puromycin (7 μg ml−1) or hygromycin (800 μg ml−1) selection for 1 week.
Cloning/lentivirus generation
CRISPR–Cas9-mediated knockout of target genes was achieved by cloning sgRNAs into pLenti-USEC or lentiCRISPRv2-puro vectors, as previously described31. In short, backbones were digested with Esp3I (New England Biosciences) and purified with a gel extraction kit (QIAGEN). sgRNAs were designed using CRISPick (Broad Institute), obtained from Integrated DNA Technologies (Coralville), annealed, phosphorylated, Esp3I-digested, and ligated into the purified digested backbones using Quick Ligase (New England Biosciences). Double sgRNA ultramers were designed and generated as previously described38. In brief, ultramers were Gibson-assembled to digested pDonor_sU6 (Addgene #69351), Esp3I-digested, and ligated to purified digested backbones. Guide RNA sequences used to make gene knockouts can be found in Supplementary Data 4. GPX4, mouse FSP1 and human FSP1 expression plasmids were generated using Gibson assembly of the respective cDNA into pLenti-v2-filler.
Lentivirus was generated by co-transfection of HEK293 cells with a viral vector and packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) using PEI transfection reagent. Cell supernatant containing lentivirus was collected 72 h after transfection and filtered through 0.45-µm PVDF filters. For in vivo experiments, lentivirus was concentrated by ultracentrifugation at 25,000 rpm for 2 h at 4 °C. The viral pellet was resuspended in PBS and stored at −80 °C until use. Viral titre was quantified with the use of a Cre-dependent GreenGo reporter cell line. For in vitro experiments, medium containing virus was collected, filtered, and added directly to recipient cells with polybrene at 8 µg ml−1 for 48 h before selection.
Mouse models
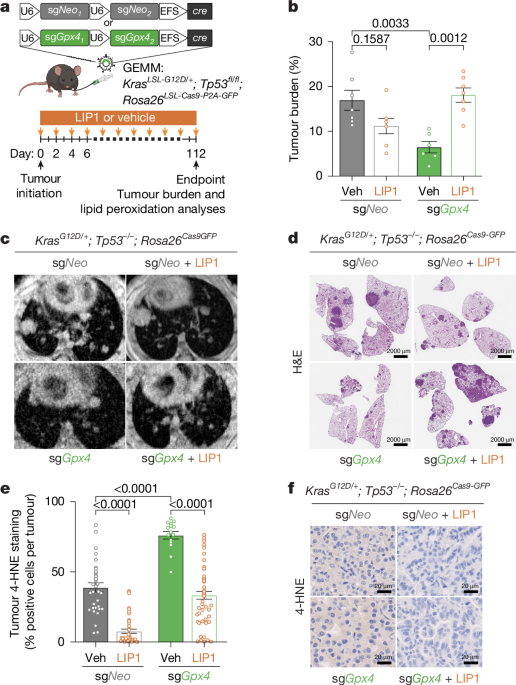
All mouse experiments described in this study were approved by the NYU Institutional Animal Care and Use Committee (IACUC). Mice were housed according to IACUC guidelines in ventilated caging in a specific pathogen-free animal facility. For all mouse studies, ≥4 mice were used for each experimental condition. KrasLSL-G12D/+; Tp53fl/fl; Rosa26LSL-Cas9/LSL-Cas9 (KPC GEMMs) mice were bred as previously described31,32,33,34,35. C57BL/6 J (JAX strain 000664) mice with the appropriate genotype, aged 8 to 12 weeks were randomly selected to begin tumour initiation studies with pUSEC lentivirus. Care was taken to ensure each experimental arm had an equal number of male and female mice. Mice were intratracheally infected with lentiviruses as described and monitored until experimental endpoint. Tumour burden was quantified by H&E staining and analysed using QuPath software as a measurement of total tumour area/total lung lobe area. All quantifications were done with investigator blinded to the respective sample genotypes. All transplantation experiments were performed using nude (JAX strain 002019), NOD SCID Gamma (NSG; JAX strain 005557 F), C57BL/6 J Fsp1-knockout (Conrad group16) or C57BL/6 J wild-type (JAX strain mice aged 8 to 12 weeks old). For mouse cell xenograft experiments, 100,000 cells in 100 µl of phosphate-buffered saline (PBS) was injected subcutaneously into each flank of the mouse. For the xenograft studies in Fig. 3 the number of cells injected per flank and whether they were injected with 50:50 PBS and Matrigel (Corning) are indicated as follows: H2009 (2 million cells + Matrigel), H1299 (1 million cells), PC9 (1 million cells), H1975 (2 million cells + Matrigel), A549 (1 million cells), 16645 (500,000 cells), KPC7 (250,000 cells). All human cell line xenograft experiments were carried out in male NSG mice unless specified. For the PDX experiment, tumours were implanted subcutaneously in male NSG mice as previously described34. Tumours were measured with callipers, and volume was calculated based on 0.5 × length × width2. The maximum tumour diameter permitted by our IACUC protocol was 2 cm, and this was not exceeded in any experiment. For orthotopic lung tumour experiments, 100,000 luciferase-expressing cells in 200 μl of PBS were injected intravenously into tail vein of male mice unless specified in the legend. Tumour growth was measured by bioluminescence (PerkinElmer IVIS Spectrum In Vivo Imaging System; D-luciferin, PerkinElmer 122799). Data were analysed using Living Image software.
Antioxidant and drug treatments
For LIP1 treatment, mice were dosed with 10 mg kg−1 LIP1 (BOC Sciences) or vehicle (2% DMSO + 40% PEG300 + 2% Tween 80 in sterile H2O) by intraperitoneal injection for frequency and duration indicated in figure schematics. For icFSP1 treatment, mice were dosed with 50 mg kg−1 icFSP1 (WuXi LabNetwork) or vehicle (45% PEG300 in sterile PBS) by intraperitoneal injection twice daily. High (TD.2108412) and low (TD.210841) irradiated vitamin E diets were obtained from Inotivco and provided ad libitum for length of time indicated in figure legends. In all experiments, mice were randomly assigned to treatment group.
Cell clonogenic and viability assays
Cell clonogenic assays were conducted by seeding 2,000 cells per well into 12-well dishes (BD/Falcon) in RPMI-1640 medium. After 12–16 h, medium containing RSL3 and/or LIP1 was added to wells. After 5 days of growth, plates were washed twice with PBS and stained with 0.5% crystal violet (Fisher Scientific) solution in 20% methanol. Plates were dried, scanned, and crystal violet was quantified by solubilization with 10% acetic acid and measurement of absorbance at 592 nm by spectrometer (Molecular Devices). Cell viability assays were conducted by seeding 2,000 cells per well into white-walled, clear-bottom, 96-well plates (Corning) in RPMI-1640 medium. After 12–16 h, medium containing RSL3, LIP1 and/or FSEN1, iFSP1 or icFSP1 was added. After three days, cell viability was assessed by CellTiter-Glo (Promega) and luminescence was measured by spectrometer (Molecular Devices).
Immunoblotting
Cells were plated to 75% confluency in a 6-well dish, and the following day cells were lysed on ice with Pierce RIPA buffer (ThermoScientific) containing 1× protease/phosphatase inhibitor cocktail (Thermo Fisher Scientific). Samples were sonicated in cooled 4 °C water (12 rounds, 30 s on and 30 s off) and then centrifuged at 14,000 rpm at 4 °C for 15 min. The supernatant was collected, and protein was quantified using the DC Rad Protein Assay kit. Protein was diluted to 1 μg μl−1 with water and 4× NuPage LDS sample buffer, then boiled at 95 °C for 10 min. Twenty micrograms of protein per well was loaded into Invitrogen 4–12% Bis-Tris gels and then transferred onto PVDF membranes using a standard protocol. PVDF membranes were then blocked using 5% BSA in TBST for 60 min at room temperature and incubated with primary antibodies in 5% BSA overnight at 4 °C. Primary antibodies were obtained as follows: GPX4 (Abcam), FSP1 (Proteintech), ACSL4 (Santa Cruz Biotechnologies), phospho-ERK (Cell Signaling), ERK (Cell Signaling) and Hsp90 (BD Bioscences). The following day, membranes were washed in TBST and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Enhanced chemiluminescent horseradish peroxidase substrate (ThermoScientific SuperSignal West PICO Plus) was added to the membrane for 1 min, and the resulting membrane was imaged using the General Electric Amersham Imager 680. For gel source data, see Supplementary Data 1.
Oxidized lipidomics
For the Gpx4-knockout experiment, cells were plated to 75% confluency in 6-well dishes. The following day, cells were treated with DMSO, RSL3 (0.5 µM), or RSL3(0.5 µM) plus LIP1(100 nM). After 8 h, cells were collected and washed in an antioxidant solution (PBS containing dibutylhydroxytoluene (100 µM) and diethylenetriamine pentaacetate (100 µM)) and centrifuged. Supernatent was discarded and cell pellets were immediately frozen in liquid nitrogen and stored at –80 °C. Frozen samples were sent on dry ice to Wayne State Lipidomics Core for metabolite extraction and LC–MS analysis.
Fsp1 wild-type and knockout tumour and cell lipidomic analysis were performed in the laboratory of Y.P.K. Fsp1 wild-type and knockout cells were plated at 75% confluency in 10 cm plates and treated as stated above. After euthanasia, mice were perfused with the antioxidant solution (+3.8% trisodium citrate), orthotopic tumours were microdissected, frozen in liquid nitrogen and stored at –80 °C. Mouse lung cancer tissues were cryopulverized and extracted with chloroform:methanol (2:1, v/v) at a tissue concentration of 25 mg mL−1. For the cells, 1.0 × 107 cells were extracted by 1 ml of chloroform:methanol (2:1, v/v). EquiSPLASH LIPIDOMIX internal standard (Avanti Polar Lipids) was added to the extraction solvent at 0.1 µg ml−1 per lipid class. Samples were sonicated on ice for 1 min using a VCX 130 probe sonicator (5 s on/off cycles), incubated for 30 min, and centrifuged at 17,000 × g for 20 min at 4 °C. The supernatant was was dried under vacuum (EZ-2 Elite, Genevac), reconstituted in 10% of the original volume with isopropanol, and transferred to glass autosampler vials for LC–MS analysis. Chromatographic separation was performed using a Waters ACQUITY UPLC CSH C18 column (100 × 2.1 mm, 1.7 µm) with a VanGuard precolumn (5 × 2.1 mm, 1.7 µm). Mobile phase A consisted of acetonitrile/water (60:40, v/v), and mobile phase B of isopropanol/acetonitrile (90:10, v/v), both containing 5 mM ammonium formate and 0.1% formic acid. The column was maintained at 65 °C with a flow rate of 0.6 ml min−1. The gradient was as follows: 0–2 min, 15–30% B; 2–2.5 min, 30–48% B; 2.5–11 min, 48–82% B; 11–11.5 min, 82–99% B; 11.5–12 min, 99% B; 12–12.1 min, 99–15% B; and 12.1–16 min, hold at 15% B for re-equilibration. Mass spectrometry was performed on a Q Exactive Plus Quadrupole-Orbitrap (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) source and operated in negative ion mode. Source settings were: sheath gas, 60 a.u.; auxiliary gas, 25 a.u.; sweep gas, 2 a.u.; spray voltage, 3.0 kV; capillary temperature, 320 °C; S-lens RF level, 50%; and auxiliary gas heater temperature, 370 °C. Parallel reaction monitoring (PRM) was carried out with the following parameters: resolution, 17,500 at m/z 200; AGC target, 2 × 105; maximum injection time, 50 ms; isolation window, m/z 1.2; and stepped normalized collision energies of 20, 30, and 40. The oxidized lipid analysis was adapted by previous PRM based analysis53. To generate the PRM inclusion list, pooled sample extracts were first analysed in DDA mode to identify the most abundant polyunsaturated phosphatidylcholine and phosphatidylethanolamine species using Lipostar254. These precursors were subjected to in silico oxidation using LPPtiger2 to predict candidate oxidized lipids. A semi-targeted DDA experiment was then conducted to confirm precursor detectability and finalize the inclusion list53. Data were analysed using Skyline (v24.1)55. Quantification was based on fragment anions derived from oxidized fatty acyl chains, with peak areas normalized to phosphatidylcholine(15:0/18:1(d7)) or phosphatidylethanolamine(15:0/18:1(d7)) internal standards from the EquiSPLASH LIPIDOMIX mixture.
CoQ9 and CoQ9H2 analysis
CoQ9 and CoQ9H2 analysis was conducted by modification of previous study56. The D9-CoQ10 standard was purchased from IsoSciences (Ambler, PA, USA). The D6-CoQ10H2 standard was synthesized from D6-CoQ10, which was obtained from Good Laboratory Practice Bioscience (Montclair, CA, USA). The CoQ9 standard was purchased from Tokyo Chemical Industry (Tokyo, Japan), and CoQ9H2 was synthesized from CoQ9. D6-CoQ10H2 and D9-CoQ10 were used as internal standards for the CoQ9H2 and CoQ9, respectively. Chloroform: methanol (2:1, v/v), containing internal standards of 0.5 µM of D6-CoQ10H2 and D6-CoQ10 was used to extract mouse lung cancer tissue at a tissue concentration of 25 mg ml−1. For the cells, 1.0 × 107 cells were extracted by 1 ml of same extraction solvent. Samples were then sonicated on ice for 1 min using a VCX 130 probe sonicator (5 s on/off cycles), incubated for 30 min, and centrifuged at 17,000g for 20 min at 4 °C. A 100 µl aliquot of the supernatant was transferred to glass autosampler vials for LC–MS/MS analysis. The liquid chromatography conditions were identical to those used for oxidized lipid analysis. Q Exactive Plus MS was operated in positive ion mode. Source settings were as follows: sheath gas, 60 a.u. (arbitrary units); auxiliary gas, 25 a.u.; sweep gas, 2 a.u.; spray voltage, 3.0 kV; capillary temperature, 320 °C; S-lens RF level, 50%; and auxiliary gas heater temperature, 370 °C. The mass range was m/z 120–1,200; resolution, 70,000 at m/z 200; AGC target, 1 × 106; and maximum injection time, 100 ms. By using EL–MAVEN (v0.12.0), the LC–MS peaks of CoQ9, CoQ9H2, D6-CoQ10H2, D6-CoQ10, and D9-CoQ10 were identified by matching with standard library and their peak areas were extracted with 10 ppm error range. Standard curves were generated using known concentrations of CoQ9H2 and CoQ9, and used to calculate the concentrations of CoQ9H2 and CoQ9 in the samples following an algorithm described in a previous study56.
Immunohistochemistry
Tumour-bearing mice were euthanized by carbon dioxide asphyxiation, after which the lungs were dissected and fixed in 4% PFA solution overnight. Fixed lungs were washed with PBS 2 times, transferred, and stored in 70% ethanol, until cassette loading and paraffin embedding. Sections were cut and stained with H&E. For immunohistochemistry with the exception of TUNEL staining, sections were immunostained on a Leica BondRX automated stainer according to the manufacturer’s instructions. In brief, tissues underwent deparaffinization online, followed by epitope retrieval for 20 min at 100° with Leica Biosystems ER2 solution (pH9, AR9640), endogenous peroxidase activity blocking with H2O2, and non-specific binding site blocking with Rodent Block M (Biocare, RBM961L) and Bond Primary Antibody Diluent (Leica Biosystems, AR9352). Sections were then incubated with primary antibodies against GPX4 (Abcam), FSP1 (obtained from M. Conrad), 4-HNE (JaICA), Ki67 and cleaved caspase-3 for 60 min at room temperature. Primary antibodies were detected with anti-rat HRP-conjugated polymer (Biocare, BRR4016H), 3,3′-diaminobenzidine (DAB) substrate (provided in the Leica BOND Polymer Refine Detection System, DS9800), and for 4-HNE staining Bond DAB Enhancer (Leica Biosystems, AR9432). Following counter-staining with haematoxylin, slides were scanned at 40× on a Hamamatzu Nanozoomer (2.0HT). TUNEL staining was performed by Histowiz according to their protocol.
For OPAL imaging, coronal 5-µm sections were immunostained on a Leica BondRx auto-stainer according to the manufacturer’s instructions. In brief, sections were deparaffiinized online and then treated with 3% H2O2 to inhibit endogenous peroxidases, followed by antigen retrieval with either ER1 (Leica, AR9961; pH6) or ER2 (Leica, AR9640; pH9) retrieval buffer at 100 °C for 20 min. After blocking with either Rodent Block M (Biocare, RBM961L) or Primary Antibody Diluent (Leica, AR93520), slides were incubated with the first primary antibody (FSP1 (obtained from M. Conrad); phospho-ERK1/2, CST; GPX4, Abcam) and secondary HRP polymer pair, followed by HRP-mediated tyramide signal amplification with a specific Opal fluorophore. Once the Opal fluorophore was covalently linked to the antigen, primary and secondary antibodies were removed with a heat retrieval step. This sequence was repeated three more times with subsequent primary and secondary antibody pairs, using a different Opal fluorophore with each primary antibody (see table below for primary antibody sequence and reagent details). After antibody staining, sections were counterstained with spectral DAPI (Akoya Biosciences, FP1490) and mounted with ProLong Gold Antifade (ThermoFisher Scientific, P36935). Semi-automated image acquisition was performed on an Akoya Vectra Polaris (PhenoImagerHT) multispectral imaging system. Slides were scanned at 20× magnification using PhenoImagerHT 2.0 software in conjunction with Phenochart 2.0 and InForm 3.0 to generate unmixed whole slide qptiff scans. All image files were uploaded to the NYUGSoM’s OMERO Plus image data management system (Glencoe Software).
Flow cytometry
Lung tissue was processed into a single-cell suspension for flow cytometry as previously described42,57. In brief, prior to euthanasia, mice were injected with 2 μg of Anti-mouse CD45-APC conjugated antibody (Biolegend, Clone 30-F11, 103111) retro-orbitally. Lungs were harvested and digested with Collagenase (Sigma-Aldrich, C5138) and deoxyribonuclease I (Sigma-Aldrich, DN25) followed by red blood cell lysis. Single cells were then resuspended in fluorescence-activated cell sorting (FACS) buffer and stained using the following antibodies: CD45 (Biolegend, 103132), CD11b (Biolegend, 101216), CD11c (Biolegend, 117324), Ly6G (Biolegend, 127622), MHCII (BD, 748708), CD103 (Biolegend, 121433), CD64 (Biolegend, 139309), SiglecF (BD, 740956), MertK (R&D, BAF591), CD45 (BD, 748371), CD3e (BD, 740854), CD4 (Invitrogen, MCD0428), CD8a (EBioscience, 563152) and Secondary (Streptavidin) (BD, 564176). Samples were run on the BD LSRFortessa and analysed on FlowJo version 10. Gating strategy is presented in Supplementary Data 2.
TCGA analyses
Gene expression profiles of primary tumours and relevant clinical data of 515 patients with LUAD were obtained from The Cancer Genome Atlas6 (TCGute.org). GPX4 and FSP1 (AIFM2) mutational status of TCGA tumour samples was retrieved from cBioPortal151 using the TCGA PanCancer Atlas collection (https://gdc.cancer.gov/about-data/publications/pancanatlas). For survival data, patients were stratified based on GPX4 or FSP1 expression and overall survival rates were plotted to compare patients with high-GPX4 or FSP1 expression (top 50% above median expression) with the rest of the cohort (n = 464 patients). All survival analyses were conducted using the survival curve analyses in GraphPad Prism v9.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism v9. All data are expressed as mean plus standard error of the mean, unless otherwise specified. Data were analysed by statistical tests as indicated in figure legends. All tests were two-tailed and replicates are biological unless otherwise stated. All western blots were replicated at least three times with results reproducible of data shown in figures. All in vitro assays were replicated at least three times with a minimum of n = 3 biological replicates per group for statistical power. For all in vivo experiments, the minimum sample size was four independent mice or tumours and respective sample size per genotype or condition is further specified in the figure legend. Sample size was not calculated but was chosen in each experiment based on previous experience with various models and to ensure that there were enough samples for statistical power. All in vivo experiments were replicated at least twice with results reproducible of data shown in figures. When representative images are shown, a minimum of three samples from the larger cohort were stained from each group. In the case of representative MRI images, all mice from the cohort were imaged. During sample processing and analysis for the lipidomic the samples were given numeric IDs which after analysis, were unblinded and graphed. For histological and immunohistochemistry analysis, researcher was blinded to the sample condition. The investigators were not blinded during most other data collection or analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.