Cell culture
Human lymphoblastoid cell lines (LCL) GM03786, GM04890 and GM03417 were obtained from Coriell. All LCL cell lines were cultured in RPMI 1640 (Gibco) with l-glutamine supplemented with 15% fetal bovine serum (FBS) in a 37 °C incubator with 5% CO2.
ONT sequencing
The ultra-high molecular weight DNA was extracted from frozen cell pellets using the NEB Monarch HMW DNA Extraction Kit for Tissue and assessed for fragment size using a pulsed field gel. The fragments span from 50 kb to 1,000 kb in size. Genomic DNA libraries were prepared using the NEB_5ml_Ultra-Long Sequencing Kit (SQK-ULK001)-promethion protocol from Oxford Nanopore. Each library was loaded onto a FLO-PRO002 flow cell and ran for 72 h with two subsequent loadings at 24-h intervals. The libraries were sequenced on a PromethION (Oxford Nanopore) running MinKNOW software v.22.12.5. Basecalling and modified base detection (5mC) were performed on-instrument using Guppy 6.4.6 with the following model: dna_r9.4.1_450bps_modbases_5mc_cg_hac_prom.cfg.
PacBio HiFi sequencing
PacBio library preparation was conducted using the SMRTBELL Prep Kit 3.0. The prepared libraries were quantified and sequenced on a PacBio Revio system with Instrument Control Software v.12.0.0.183503 and chemistry v.12.0.0.172289. Sequencing was performed using two SMRT Cells, each with a movie length of 24 h.
Using Pacific Biosciences SMRTbell Prep Kit 3.0 with binding kit 102-739-100 and sequencing kit 102-118-800, three libraries (one per sample). The Megarupter (Diagenode) was used for shearing and SageELF (Sage Science) was used for size selection. Library size was assessed using a FemtoPulse (Agilent). Each library was run on v.25M SMRT Cells using the first generation polymerase and chemistry v.1 (P1-C1). Sequencing was performed on a PacBio Revio system running instrument control software v.12.0.0.183503 and a movie collection time of 24 h per SMRTCell. Using PacBio SMRTLink v.12.0.0.172289, CCS/HiFi reads generated on-instrument using ccs v.7.0.0, lima v.2.7.1 (demultiplexing), and primrose v.1.4.0 (5mC calling).
Hi-C sequencing
Hi-C libraries were generated according to manufacturer’s directions using the Arima High Coverage Hi-C User Guide for Mammalian Cell Lines (A160161 v.01) and Arima-HiC+ User Guide for Library Preparation Using the Arima Library Prep Module (A160432 v.01). Starting with 5 million cells per sample, the Standard Input Crosslinking protocol was followed, resulting in 1.49–1.86 μg of DNA available per sample to generate large proximally ligated DNA as assessed using the Qubit Fluorometer (Life Technologies). Library preparation was performed using the S220 Focused-ultrasonicator (Covaris) to shear samples to 550 bp followed by a DNA purification bead cleanup with no size selection, and 5 or 7 cycles of library PCR amplification per sample. Resulting short fragment libraries were checked for quality and quantity using the Bioanalyzer (Agilent) and Qubit Fluorometer (Life Technologies). Libraries were pooled, requantified and sequenced as 150 bp paired reads on both the Illumina NextSeq 2000 and NextSeq 500 instruments to obtain at least 600 M read pairs per sample, using real-time analysis and instrument software versions current at the time of processing. Demultiplexing was performed with bcl-convert v.3.10.5. The cut sites (^) for the enzymes used were ^GATC, G^ANTC, C^TNAG and T^TAA.
Library construction for GM04890 and GM03786
Libraries were generated from 100 ng genomic DNA using Covaris LE220 plus to shear the DNA and the 2S Plus DNA Library Kit (Integrated DNA Technologies 10009878) for library preparation. To minimize coverage bias, only four cycles of PCR amplification were used. The median insert sizes were approximately 300 bp. Libraries were tagged with unique dual index DNA barcodes to allow pooling of libraries and minimize the impact of barcode hopping. Libraries were pooled for sequencing on the NovaSeq X plus (Illumina) across 14 lanes to obtain at least 369 million 151-base read pairs per individual library.
Library construction for GM03417
PCR-free libraries were generated from 1 μg genomic DNA using a Covaris R230 to shear the DNA and the TruSeq DNA PCR-Free HT Sample Preparation Kit (Illumina) for library preparation. The median insert sizes were approximately 400 bp. Libraries were tagged with unique dual index DNA barcodes to allow pooling of libraries and minimize the impact of barcode hopping. Libraries were pooled for sequencing on the NovaSeq X plus (Illumina) across 7 lanes on 25B flowcells to obtain at least 388 million 151-base read pairs per individual library.
Assembly methods
Phased genome assemblies were generated using Verkko (v.1.4.1)23. The assembly process integrated PacBio HiFi reads and Oxford Nanopore (ONT) reads, with Hi-C reads used specifically for the phasing. The ONT reads included ultra-long reads, defined as reads that are at least 100 kb in length. Verkko was run with the command:
samples = (″GM03417″ ″GM03786″ ″GM04890″)
for sample in ″$samples[@]″; do
verkko –slurm -d $sample \
–screen human \
–graphaligner conda/bin/GraphAligner \
–mbg conda/bin/MBG \
–hifi-coverage 30 \
–hifi $sample/HiFi/*fa.gz \
–nano $sample/ONT/fastq/*fq.gz \
–hic1 $sample/HiC/*_1_[ACTG]*.fastq.gz \
–hic2 $sample/GM03786/HiC/*_2_[ACGT]*.fastq.gz
done
Haplotype-consistent contigs and scaffolds were automatically extracted from the labelled Verkko graph, with unresolved gap sizes estimated directly from the graph structure. After the assembly was generated, we collapsed all nodes composed of only rDNA k-mers into a single node and added telomere nodes to the graph to indicate ends of chromosomes using the commands:
seqtk hpc rDNA.fasta > rDNA_compressed.fasta
seqtk telo assembly.fasta > assembly.telomere.bed
mash sketch -i 8-hicPipeline/unitigs.hpc.fasta -o compressed.sketch.msh
$mash screen compressed.sketch.msh rDNA_compressed.fasta | awk ‘if ($1 > 0.9 & & $4 < 0.05) print $NF’ > target.screennodes.out
python remove_nodes_add_telomere.py -r target.screennodes.out -t assembly.telomere.bed
In this simplified graph, the Robertsonian translocation was apparent in all cases (Extended Data Figs. 1–3). We extracted the assembly path corresponding to the ROB and identified gaps in the assembly. There was one gap in GM03417, one gap in GM03786 and two gaps in GM04890. Manual interventions were used to complete the chromosomes.
Assembly quality evaluation
We evaluated the quality and gene completeness of the genome assemblies using two approaches: a k-mer-based, reference-free method and a gene content assessment. For the k-mer-based evaluation, we employed Merqury55, a tool that assesses assembly completeness and accuracy without relying on a reference genome. Merqury uses k-mer frequencies from sequencing reads to estimate the quality value of the assemblies, which represents the phred-scaled error rate. For our evaluation, we used PacBio HiFi reads for the quality value estimation.
To assess gene completeness, we used compleasm56, a tool based on BUSCO. Compleasm evaluates the presence and integrity of a curated set of BUSCOs expected to be present in the genomes of the taxonomic group under study. We used the primate-specific BUSCO dataset, which includes 13,780 genes, to quantify the completeness, duplication and fragmentation of conserved genes in our assemblies.
PRDM9 site predictions and density
In 147 human haploid genomes (from 72 diploid individuals plus the haploid CHM13 and diploid HG002 genomes), predicted PRDM9 DNA binding sites were identified by using Motifence (v.0.1.1, commit fb1ebc0; https://github.com/AndreaGuarracino/motifence) to find DNA sequences matching the canonical 13-mer motif CCNCCNTNNCCNC57 or its reverse complement. To compute the density of PRDM9 DNA binding sites per kb in SST1 regions, SST1 arrays were first identified using TideHunter58. For a region to be defined as an SST1 array, the following criteria were applied: monomeric unit within the array had to be at least 500 bp in length, there had to be at least two monomers, and the monomers had to overlap with RepeatMasker (v.4.1.5, http://repeatmasker.org/) SST1 annotations. The PRDM9 density was then calculated by dividing the number of PRDM9 binding sites in the SST1 regions by the total length of these SST1 regions. PRDM9 alleles were found by conducting a BLAST search (blast-plus/2.13.0) on GM3417, GM3786 and GM4890 with the A allele as the reference. To identify genotypes, these hits were aligned to the 69 alleles from Alleva et al.30 using MUSCLE and visualized in Geneious Prime 2024.0.7.
In the chimpanzee genome, PRDM9 site density in sites per kb on SST1 regions was calculated using R and Bioconductor. The function vmatchPattern from the Biostrings library was used to map the occurrence of the chimpanzee PRDM9 motifs: prdm9_E CNNCCNAANAA, prdm9_W CNGNNAANANTT and prdm9_pt1 ANTTNNATCNTCC, or their reverse compliments, on the genome. SST1-containing regions were then queried for overlap of PRDM9 sites using the countOverlaps function from the GenomicRanges library. Query width was used to calculate sites per kb. SST1 regions larger than 10 kb were broken into 3-kb tiles to approximate resolution near SST1 feature size. Background PRDM9 site density was assessed in two ways. Random background PRDM9 density for each chromosome was determined using 100 randomly chosen 3-kb segments. To account for GC bias, the genome was scored for GC content at 3 kb resolution, and fragments within one s.d. of the average GC content of the SST1-containing elements were chosen to calculate background PRDM9 site density.
SST1–segmental duplication association
To examine associations between SST1 repeats and segmental duplications, we performed the following analysis in 147 human genomes (from 72 diploid individuals plus the haploid CHM13 and diploid HG002 genomes). First, repetitive regions in the genomic sequences were masked using RepeatMasker (v.4.1.5, http://repeatmasker.org/) and Tandem Repeats Finder (v.4.09.1)59. Segmental duplications were then identified using SEDEF (v.1.1)60 on each haploid masked genome. SST1 repeats were detected using RepeatMasker and refined with TideHunter, as described above. Finally, we used the R package regioneR (v.1.36.0)61 to perform permutation testing (n = 10,000) to assess the significance of spatial associations between SST1 repeats and segmental duplications. This analysis was conducted on 147 haplotype-resolved genomes to provide a comprehensive view of these genomic features across diverse human genomes.
SST1 monomer characterization
We used RepeatMasker to find the regions. We retrieved all fasta files with 1 kb of flanking regions for all arrays. Then, we manually curated all clusters using visual inspection by generating dot plots with the Dotlet applet62 with a 15 bp word size and 60% similarity cut-off. We made regressive changes in the consensus sequences used and that enabled us to describe the sequences properly. By manual curation, we were able to identify the beginning and end of the arrays and each monomer regarding the consensus generated. All monomeric sequences analysed were characterized with the same initial and final point regarding the consensus for the sake of alignment.
Maximum-likelihood phylogenetic analysis
We aligned all SST1 full-length monomeric sequences retrieved from assembled genomes using MUSCLE63. We conducted the phylogenetic analysis by using the maximum-likelihood method based on the best-fit substitution model (Kimura two-parameter + G, parameter = 5.5047) inferred by Jmodeltest2 with 1,000 bootstrap replicates. Bootstrap values higher than 75 are indicated at the base of each node.
Chromosome spreads, FISH and immunoFISH
For the preparation of chromosome spreads, cells were blocked in mitosis by the addition of Karyomax colcemid solution (0.1 µg ml−1, Life Technologies) for 6–7 h. Adherent fibroblast cells were collected by trypsinization. Collected cells were incubated in hypotonic 0.4% KCl solution for 12 min and pre-fixed by addition of methanol:acetic acid (3:1) fixative solution (1% total volume). Pre-fixed cells were spun down and then fixed in methanol:acetic acid (3:1).
For SST1 and centromere FISH, spreads were dropped on a glass slide and incubated at 65 °C overnight. Before hybridization, slides were treated with 0.1 mg ml−1 RNAse A (Qiagen) in 2× SSC for 45 min at 37 °C and dehydrated in a 70%, 80% and 100% ethanol series for 2 min each. Slides were denatured in 70% deionized formamide/2× SSC solution pre-heated to 72 °C for 1.5 min. Denaturation was stopped by immersing slides in 70%, 80% and 100% ethanol series chilled to −20 °C. Labelled DNA probes were denatured separately in a hybridization buffer by heating to 80 °C for 10 min before applying to denatured slides. Fluorescently labelled human centromere probes for D13Z1/D21Z1 and D14Z1/D22Z1 were from Cytocell. The biotin-labelled BAC probe for SST1 (RP11-614F17) was obtained from Empire genomics. Specimens were hybridized to the probes under a glass coverslip or HybriSlip hybridization cover (GRACE Biolabs) sealed with the rubber cement or Cytobond (SciGene) in a humidified chamber at 37 °C for 48–72 h. After hybridization, slides were washed in 50% formamide/2× SSC 3 times for 5 min per wash at 45 °C, then in 1× SSC solution at 45 °C for 5 min twice and at room temperature once. For biotin detection, slides were incubated with fluorescent streptavidin conjugated with Cy5 (ThermoFisher Scientific) for 2–3 h in PBS containing 0.1% Triton X-100 and 5% bovine serum albumin (BSA), and then washed 3 times for 5 min with PBS/0.1% Triton X-100. Slides were mounted in Vectashield containing DAPI (Vector Laboratories). Confocal z-stack images were acquired on the Nikon TiE microscope equipped with PlanApo 100× oil immersion objective NA 1.45, Yokogawa CSU-W1 spinning disk, Flash 4.0 sCMOS camera (Hamamatsu), and NIS Elements software.
For chimpanzee and bonobo cell lines, chromosome spreads specimens were hybridized to the probes under a glass coverslip or HybriSlip hybridization cover (GRACE Biolabs) sealed with rubber cement or Cytobond (SciGene) in a humidified chamber at 37 °C for 48 h. After hybridization, slides were washed in 50% formamide/2× SSC 3 times for 5 min per wash at 45 °C, then in 1× SSC solution at 45 °C for 5 min twice and at room temperature once. For biotin detection, slides were incubated with fluorescent streptavidin conjugated with 488 (ThermoFisher Scientific) for 45 min in PBS containing 0.1% Triton X-100 and 5% BSA, and then washed 3 times for 5 min with PBS/0.1% Triton X-100. Slides were mounted in Vectashield containing DAPI (Vector Laboratories). Confocal z-stack images were acquired on the Zeiss LSM 800 microscope equipped with a 63×/1.4 Plan-Apochromat 63× oil immersion objective and Zen Blue software.
For chimpanzee and bonobo, we used the following SST1-sf1 probe: (5′-AGGCCAAATATCAGCTGCAAATTCAATCATCCATCAGCCCTCTGCCTACCTCTTCCTTTGAAAGGGCAGTGGCCGGCCCGGCTTGTAAAAGCCCTGGGGTTCCAGAAAGCCGACCGCGCTTTACAGAACAACTGTAATGAGGAACACAGGCGAATCCGAGGGGGTGACCATGTGACCACGCGTGGTACTGGCCAATCCCACAGCAGCTGGTGTTAATGTGTGTCACCGGAGGCATACGGGGCGACGGCGAAACAAAGGGTGGTGTCCAGGAATGTGCCGGTGGATGGGGAAACGGGTGACCTTTCCATCAATGCCAACGAAAATCAAAGAACAACTGGGACCCGGGGGTTGGGGGTGCCGCCTGTGCCTGACCCAAGCCACGTTTTCAAATGCCTACCAGAGGAGCAGAGAGGTTTCTGCAAAATTCGCAGCATCCCCAATCCTCCACCGACCTGGTAGCCCTGACGAAACTTCGGCTGGCACAAACCCAGAGAGGGTGGGGAGTCATACAGCAGAGGAGAGCAGCCCAGGGGCACGCAGGCCGACCCGTCATCGAGATCACGGACGGCCGCACGACTTTTCGGGAGACTCACCCCAGCCAACACCGTCCGTGCAGGCCTGAGGCTGGTATCCCGTGCTGCTTCCCCCCGTCTCCGCCTGGGGTTTCCTCATCAAGGTCGGCCCTTTGCGACTCCTGGCATCCGGAGACGTTCCCTTCGACCCCGTGGAGAGGTGAGGCTTTAGCCTCAGAGCCTCGACACCCAAGCACTGCAACGGAGGGCTCCTGCTCTGCCAAGCCTCGGGGCCTGGTTTCTAAGAAAACCGTGGGAACCACTGTGACGGGAGATACCGCTCGCGCCTCGCGCATGCGCATTGGCCGAGCCGATTCGCGCTCCACTGCTGACAGATAGGCTGCGTCCGCTTTAAATATCGCCACCACCACGCGGCGGCCTTGGTGCTCCTGCTGCCGCTGCGGCGGCGGCTGGATCCTGGGTCCTGTTTGGGGCGGCATGCGAAAGGGGACCGCGGGTGTCTCGTCCTGTCCCAGGCCCACACCCCCAGGGGTCCTGTCCACAGGACCTGCTTCAGCCGACTTCCACCGAGGGAGGGGGAGCTTCAGGACGCCTGCTGTGTTCTCCGGACTCCCGTTGAGATCCGATTTTGGCCCTCTCCGAGTGAGATAGGACGAGCTCACCACACCCGGACAGGCCGGCAGGGCCTC GCTGCAGCACAGAATGATCCCGTAGGTCTGA-3′).
For CENP-B and CENP-C immunoFISH, freshly prepared chromosome spreads were dropped on a glass slide, washed with PBS/0.1% Triton X-100, and blocked with 5% BSA in PBS/0.1% Triton X-100. Primary antibody (rabbit polyclonal anti-CENP-B, Abcam, ab25734, rabbit polyclonal anti-CENP-C, Millipore, ABE1957) and secondary antibody (goat anti-rabbit Alexa Fluor 647, ThermoFisher Scientific) were diluted in 5% BSA/PBS/0.1% Triton X-100. Specimens were incubated with primary antibody overnight, washed 3 times for 5 min, incubated with secondary antibody for 2–4 h and washed again 3 times for 5 min. All washes were performed with PBS/0.1% Triton X-100. After antibody incubation, spreads were post-fixed in 2% paraformaldehyde diluted in PBS for 15 min, washed in PBS, and processed for FISH as described above, starting with an ethanol dehydration series. DNA was stained with 1.5 µg ml−1 DAPI. Confocal z-stack images of CENP-B immunoFISH were acquired on the Nikon TiE microscope as described above. For SIM performed on CENP-C immunoFISH, slides were rinsed in ddH2O, air-dried in the dark, mounted in ProLong Glass antifade mountant (ThermoFisher Scientific) and allowed to cure for at least 24 h before imaging. z-stack images were acquired on an Elyra 7 Lattice SIM2 microscope (Zeiss) equipped with two PCO.edge 4.2 sCMOS cameras, four high power continuous wave lasers (405, 488, 561 and 642 nm) and a Zeiss PlanApo 63× oil immersion objective NA 1.4. The illumination pattern was set to 15 phases, and the z-stack spacing was set at 100 nm. Raw SIM images were reconstructed using the ZEN Black software (Zeiss) with 10.5 manual adjustments for sharpness and best-fit settings for all channels except 405 nm (DAPI), which was processed in the widefield mode. Image pre-processing for SPA-SIM included channel alignment; this analysis randomizes any residual chromatic shifts by averaging randomly oriented chromosomes.
For CENP-A and NDC80 immunoFISH experiments, fibroblasts plated on 150-mm dishes were treated with 100 µM Monastrol (Tocris Bioscience) and 100 µM Apcin (Selleck Chemicals) for 5 h and collected by mitotic shake-off. Collected cells were further incubated with Karyomax colcemid solution (0.1 µg ml−1, Life Technologies) for 15 min. After that, cells were spun down and resuspended in 0.075 M KCl swelling buffer containing 10 mM HEPES, incubated at room temperature for 12 min, washed with ice-cold PBS and kept on ice. Cells (3–4 × 105) were spun onto glass slides using Shandon Cytospin 4 centrifuge (Thermo scientific) at 700–1000 rpm for 3–5 min, washed in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl, pH8, 0.5 mM EDTA, 0.1% (v/v) Triton X-100) and blocked in 5% (w/v) BSA/KCM for 30 min. Slides were then incubated with primary antibodies (mouse anti-CENP-A (3-19) Enzo ADI-KAM-CC006, or mouse anti-NDC80 (9G3.23) ThermoFisher Scientific MA1-23308 with rabbit anti-CENP-A ProSci 30-143) used at 1:100 dilution for 1 h at room temperature, washed 3 times in KCM for 5 min, followed by incubation for 1 h with species-specific secondary antibodies conjugated with Alexa Fluor Plus dyes at 2 µg ml−1, washed again and post-fixed in in 4% (v/v) paraformaldehyde/KCM for 10 min. Fixed slides were incubated in 50% glycerol/PBS at 4 °C for at least 1 h or overnight. Before hybridization, slides were subjected to a freeze-thaw treatment by dipping into liquid nitrogen, then treated with 0.1 N HCl for 5 min, washed twice in 2× SSC buffer, and pre-incubated in 50% formamide/2× SSC overnight. Fluorescently labelled probes were pre-denatured for 7 min at 80 °C, followed by incubation with the specimen for 3 min at 74 °C, and hybridized under HybriSlip hybridization cover (GRACE Biolabs) sealed with Cytobond (SciGene) in a humidified chamber at 37 °C for 24–48 h. After hybridization, slides were washed in 50% formamide/2× SSC 3 times for 5 min per wash at 45 °C, then in 1× SSC solution at 45 °C for 5 min twice and at room temperature once. DNA was stained with 1.5 µg ml−1 DAPI. After staining was completed, slides were rinsed in ddH2O, air-dried in the dark, mounted in ProLong Glass (ThermoFisher Scientific) and allowed to cure for at least 24 h before SIM imaging.
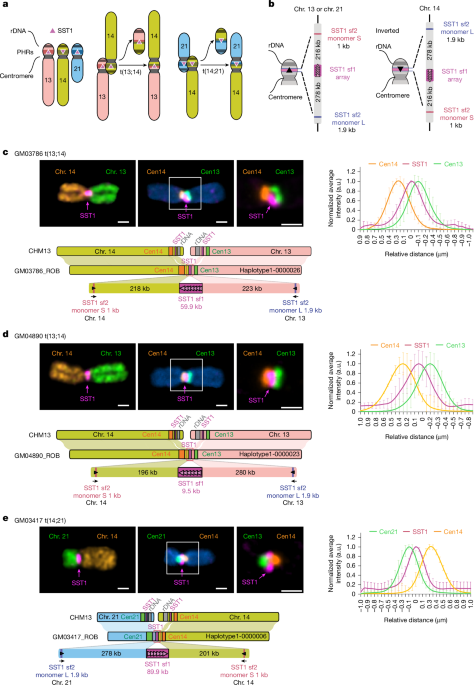
Centromere intensity profiling of centromere and SST1 FISH and CENP-B immunoFISH
Maximum intensity projections from spinning disk confocal z-stacks were generated, and chromosomes of interest were segmented manually on the basis of DNA and centromere labelling. Segmented chromosomes from each cell line were oriented vertically and assembled in a new stack consisting of identified specific chromosomes from multiple chromosome spreads. Intensity plot profiles were generated from 2 µm vertical lines with the width of 10 pixels drawn through centromeric regions of each chromosome. Intensity profiles were combined by channel, fit to single Gaussian functions, and aligned to the peak of the Gaussian of the indicated channel. These profiles were then averaged together and normalized to the maximum intensity of each peak. For each chromosome from each cell line, at least ten intensity profiles were averaged and plotted with the s.d. All image processing and analysis were performed using ImageJ/FIJI. A detailed description of this type of analysis and relevant plugins are available at https://research.stowers.org/imagejplugins/spasim.html.
Semi-automated intensity profiling of CENP-C immunoFISH from SIM images
Reconstructed SIM images were mean projected, except for the DAPI channel, which had the slice of highest contrast selected. ROBs and corresponding normal acrocentric chromosomes were identified using centromere FISH signals and segmented manually or with a Cellpose model trained on a combination of the DAPI and centromere signals. Individual chromosomes were transferred to a new image and oriented vertically using a second Cellpose model trained to find a skeleton of the chromosome. Bent chromosomes were straightened in ImageJ/FIJI by manually drawing two annotation lines across centromeres, one through each kinetochore. The straightened images were then aligned to the peak of the specified centromere FISH signal used as the anchor point, and the line intensity profiles were aggregated over multiple images and split by cell line. At least ten chromosomes were analysed for each instance from each cell line. All analysis was performed in ImageJ/FIJI and Python with code at https://github.com/jouyun/Gerton_Robertsonian_2024.
Methylation calls
HiFi BAM and ONT FASTQ files with 5mC methylation calls as MM and ML tags were aligned against the generated assemblies using pbmm2 (v.1.13.0, https://github.com/PacificBiosciences/pbmm2) for HiFi reads and Winnomap (v.2.03)64, for ONT reads. The alignments were then converted to sorted BAM files containing only primary mappings with samtools (v.1.17)65:
# HiFi reads
pbmm2 align genome.mmi bam_with_meth_calls -j 42 > output.bam
samtools view -@ 24 -Sb -F 2048 output.bam | samtools sort -@ 24 -T temporary_directory – > output.bam
samtools index output.bam
# ONT reads
winnowmap -t 48 -W genome_repetitive_k15.txt -ax map-ont -y assembly_fasta fastq_with_meth_calls > output.sam
samtools view -@ 24 -Sb -F 2048 output.sam | samtools sort -@ 24 -T temporary_directory – > output.bam
samtools index output.bam
Aggregated methylation percentages at all CpGs were obtained using modbam2bed (v.0.10.0, https://github.com/epi2me-labs/modbam2bed) with bases with >0.8 probability called “methylated” and bases with <0.2 probability called “unmethylated”:
modbam2bed -t 48 -e -m 5mC –cpg -a 0.20 -b 0.80 assembly_fasta output.bam > output.bed
CUT&RUN library preparation
The CUT&RUN assay was performed using the CUT&RUN Assay Kit (86652, Cell Signaling Technology) in accordance with the manufacturer’s protocol. For each condition, 250,000 cells were pelleted and washed in 1× wash buffer, prepared from 10× wash buffer (31415, Cell Signaling Technology), 100× spermidine (27287, Cell Signaling Technology) and 200× protease inhibitor cocktail (7012, Cell Signaling Technology). Cell suspensions were then incubated with concanavalin A-coated beads for 5 min at room temperature to facilitate binding, followed by resuspension in 1× binding buffer containing 100× spermidine, 200× protease inhibitor cocktail, 40× digitonin solution (Cell Signaling Technology, 16359) and antibody binding buffer (Cell Signaling Technology, 15338). For the detection of CENP-A–DNA interactions, a monoclonal antibody against CENP-A (Enzo, ADI-KAM-CC006-E) was employed at a 1:50 dilution. As controls, tri-methyl-histone H3 (Lys4) (Cell Signaling Technology, 9751, C42D8 rabbit monoclonal antibody) was used at 1:50 dilution as a positive control, while a rabbit IgG XP isotype control (Cell Signaling Technology, 66362, DA1E monoclonal antibody) was applied at 1:10 dilution as a negative control. Antibody incubation was conducted at 4 °C overnight (16 h). Later, the beads were subjected to magnetic separation and washed in digitonin buffer.
The beads were then resuspended in digitonin buffer containing pAG-MNase enzyme (40366) and incubated at 4 °C for 1 h. Following another wash in digitonin buffer, the beads were treated with calcium chloride in digitonin buffer and incubated at 4 °C for 30 min to facilitate MNase activation. The enzymatic digestion was terminated by adding 1× stop buffer (prepared from 4× stop buffer (Cell Signaling Technology, 48105), digitonin solution and 200× RNase A (Cell Signaling Technology, 7013)). For normalization, spike-in DNA (Cell Signaling Technology, 40366) was introduced at a final concentration of 10 pg μl−1 (1:100 dilution). Samples were then incubated at 37 °C for 10 min, and the supernatants were collected by centrifugation. DNA was liberated via incubation at 65 °C for 2 h before purification. Input chromatin samples were sheared to fragments ranging from 100–700 base pairs using a Covaris S2 sonicator prior to purification.
DNA purification was performed using a DNA purification with spin columns kit (Cell Signaling Technology, 14209). DNA concentration was assessed using the Qubit dsDNA HS kit for the Qubit Fluorometer.
CUT&Tag library preparation
For anti-CENP-A CUT&Tag, library preparation was used the CUT&Tag-IT kit from Active Motif (53160). Each experiment was performed for 500,000 fresh cells. Fresh cells were washed using 1× wash buffer and nuclei were isolated and incubated with activated concanavalin A-coated magnetic beads in 2 ml PCR tubes at room temperature for 10 min. A 1:100 dilution of primary antibody anti-CENP-A (human) monoclonal antibody (D115-3) in antibody buffer was added and nuclei were incubated overnight at 4 °C. The next day tubes were incubated on a magnetic tube holder and supernatants were discarded. Secondary antibody (rabbit anti-mouse) was diluted at 1:100 in Dig-Wash buffer and nuclei were incubated for 1 h on an orbital rotator at room temperature. Nuclei were washed three times in Dig-Wash buffer and then incubated with a 1:100 dilution of CUT&Tag-IT pA–Tn5 Transposomes for 1 h on an orbital rotator at room temperature. After, 125 μl of tagmentation buffer was added to each sample. To stop tagmentation, 4.2 μl 0.5 M EDTA, 1.25 μl 10% SDS and 1.1 μl 10 mg ml−1 proteinase K was added to each reaction and incubated at 55 °C for 1 h. DNA was barcoded and amplified using the following conditions: a PCR mix of 25 μl NEBNext 2× mix, 2 μl each of barcoded forward and reverse 10 μM primers, and 21 μl of extracted DNA was amplified at: 58 °C for 5 min, 72 °C for 5 min, 98 °C for 45 s, 16× 98 °C for 15 s followed by 63 °C for 10 s, 72 °C for 1 min. Amplified DNA libraries were purified by adding a 1.1× volume of SPRI beads to each sample and incubating for 10 min at 23 °C. Samples were placed on a magnet and liquid was removed. Beads were rinsed twice with 80% ethanol, and DNA was eluted with 20 μl elution buffer. All individually i7-barcoded libraries were mixed at equimolar proportions for sequencing.
CUT&Tag and CUT&RUN libraries and sequencing
Libraries were quantified and individually converted to process on the Singular Genomics G4 with the SG Library Compatibility Kit (700141), following the Adapting Libraries for the G4–Retaining Original Indices protocol. The converted libraries were sequenced in individual lanes on an F3 flow cell (700125) on the G4 instrument, using Instrument Control Software 23.08.1-1 with 100 bp paired reads. Following sequencing, sgdemux 1.2.0 was run to generate FASTQ files.
CUT&Tag and CUT&RUN bioinformatic analysis
CUT&Tag and CUT&RUN sequencing reads were trimmed using the trim-galore tool (v.0.6.10, https://github.com/FelixKrueger/TrimGalore), which included adapter removal. The trimmed reads of each sample were then aligned to the corresponding generated de novo assemblies using bowtie2 (v.2.5.3)66. Post-alignment, the reads were sorted and indexed using samtools (v.1.17)65, to then extract depth information for primary alignments with mosdepth67.
Pairwise sequence identity heat maps
To generate pairwise sequence identity heat maps of each centromeric region, we used a modified version of StainedGlass (v.0.6)39 with the following parameters: window=5000, mm_f = 30000, and mm_s = 1000. Our modifications were applied to visualize the identity heat maps with methylation and CENP-A CUT&Tag information included at the bottom.
Synteny plots
To visualize the alignment between the generated assemblies and the CHM13 genome, we used NGenomeSyn26 to generate the synteny plots, which were then manually curated.
Hi-C data analysis
We mapped Hi-C reads against the CHM13 genome and the phased genome assemblies of the three cell lines with the BWA aligner68, configured to handle the chimeric nature of Hi-C reads by allowing local mapping and tuning the parameters to minimize gaps. Following read mapping, for each cell line, we constructed three Hi-C contact matrices, one against CHM13 and two against the 2 haplotypes of the respective assembly, by specifying a bin size of 10,000 bp and incorporating restriction site information using HiCExplorer tools69. The resulting matrices were then binned at various resolutions (100 kb, 200 kb and 500 kb) and corrected to normalize the contact frequencies across bins and remove GC and open chromatin biases. Finally, we visualized the corrected matrices using hicPlotMatrix, applying log transformation to handle the wide range of contact counts.
Genome versions used
We leveraged multiple reference genomes and assemblies. The primary reference was T2T-CHM13v2.0. We also incorporated the recent diploid T2T-HG002v1.1 genome and 72 samples from the Human Pangenome Reference Consortium (HPRC). The HPRC samples were assembled using Verkko v.2.123, using a combination of sequencing technologies for each sample. The assembly process utilized PacBio High-Fidelity (HiFi) reads and Oxford Nanopore Technology (ONT) long reads. For phasing, we primarily used short Illumina reads. In cases where trio information was unavailable, Hi-C reads were used for phasing instead.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.