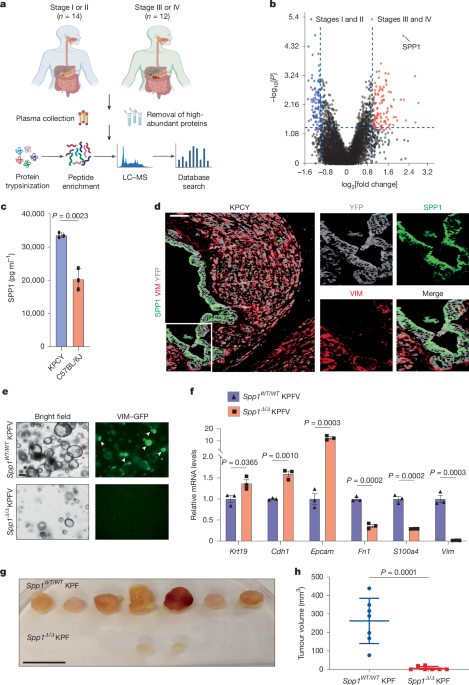
Blood samples from patients with pancreatic cancer
Peripheral venous blood samples were collected from 26 patients with pancreatic cancer at the Seventh Affiliated Hospital of Sun Yat-sen University. Plasma was isolated from the collected blood samples, and plasma proteomic analysis was subsequently performed. Informed consent was obtained from all participants, and approvals were secured from the ethics board of the Seventh Affiliated Hospital of Sun Yat-sen University for the use of these specimens in research. All clinical samples were collected with informed consent, and all experiments adhered to the regulations and approvals of the internal review and ethics board at the Seventh Affiliated Hospital of Sun Yat-sen University. The Institutional Review Board approval number is KY-2024-249-01. Clinical information on the patients, including age, sex, TNM stage, histology and differentiation, was obtained from medical and follow-up records (Supplementary Table 1).
Generation of patient-derived organoids
Human pancreatic tissue samples were obtained from patients at the Seventh Affiliated Hospital of Sun Yat-sen University who underwent surgery for pancreatic neoplasia. Informed consent was obtained from all participants, and approvals were secured from the ethics board of the Seventh Affiliated Hospital of Sun Yat-sen University for the use of these specimens in research. The tissue samples were collected in Advanced DMEM/F12 base medium with 100 µg ml–1 Primocin and minced into 1 mm2 pieces with a scissor. Minced tissue was then washed in the base medium and resuspended in 10 ml digestion medium containing Advanced DMEM/F12, 5 mg ml–1 collagenase XI, 2 mg ml–1 dispase II and 10 µg ml–1 DNase I. Tumour fragments were digested for 2–4 h on a Thermo Scientific tube rotator at 37 °C at speed 25. Digestion was stopped by adding the base medium. Tissue was then washed and centrifuged twice with the base medium and digested for 10 min with TrypLE Express (Gibco) supplemented with Y-27632 (10 µM). Tissue was then filtered through a 100 µm cell filter (Falcon), washed and centrifuged twice in the base medium. The pellet was resuspended in 5 ml red cell lysis buffer (Corning) for 5 min. Cells were then washed and centrifuged a further 2 times in the base medium, resuspended in 50 µl Matrigel per well (Corning) and plated onto pre-warmed 24-well plates. Next, 500 µl medium per well was added 15–30 min later once the Matrigel had solidified. The medium contained 1× B27 supplement, 10 mM nicotinamide, 1.25mM N-acetylcysteine, 10 nM gastrin, 100 ng ml–1 human FGF10, 100 ng ml–1 mouse Noggin, 500 nM A83-01, 100 µg ml–1 Primocin, 50 ng ml–1 EGF, 10 µM Y-27632, 10% (v/v) R-Spondin 1 conditioned medium and 50% (v/v) WNT3A conditioned medium.
Plasma proteomics
For protein extraction, cellular debris from the plasma samples was removed through centrifugation at 12,000g for 10 min at 4 °C. The supernatant was then carefully transferred to a fresh centrifuge tube, and the protein was quantified using a BCA kit (Bio-Rad) according to the manufacturer’s protocol.
For trypsin digestion, high-abundant proteins were first removed from the plasma samples using a kit (A36369, Thermo), and the solution was then treated with 5 mM dithiothreitol for 30 min at 56 °C, followed by alkylation with 11 mM iodoacetamide for 15 min at room temperature in the dark. Next, 70 μl enzyme digestion buffer was added to the beads and incubated at 95 °C for 10 min, followed by allowing the samples to return to room temperature. Trypsin was then added to a final concentration of 20 ng μl–1 for overnight digestion. The resulting peptides were desalted using C18 ZipTips (Millipore) according to the manufacturer’s instructions and then dried for subsequent mass spectrometry (MS) analysis.
For liquid chromatography–MS/MS analysis, the tryptic peptides were resuspended in solvent A and directly loaded onto a self-fabricated reversed-phase analytical column (15 cm in length, 100 μm internal diameter). The mobile phase was composed of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid and 80% acetonitrile in water). Peptides were separated using the following gradient: 0–1.6 min, 4–22.5% B; 1.6–2.0 min, 22.5–35% B; 2.0–2.6 min, 35–55% B; 2.6–2.7 min, 55–99% B; 2.7–6.8 min, 99% B; 6.8–7.6 min, 99% B, all at a constant flow rate of 300 nl min–1 on a Vanquish Neo UPLC system (ThermoFisher Scientific).
The separated peptides were analysed using an Orbitrap Astral mass spectrometer equipped with a nano-electrospray ion source. The electrospray voltage was set to 1,900 V. Precursors were analysed in the Orbitrap detector, whereas fragments were analysed in the Astral detector. The full MS scan resolution was set to 240,000 with a scan range of 380–980 m/z. The MS/MS scan was fixed with a first mass of 150.0 m/z at a resolution of 80,000. Higher-energy collisional dissociation fragmentation was performed at a normalized collision energy of 25%. The automatic gain control target was set at 500%, with a maximum injection time of 3 ms.
For database searches, the data-independent acquisition (DIA) data were processed using the DIA-NN search engine (v.1.8). Tandem mass spectra were searched against the Homo_sapiens_9606_SP_20231220.fasta database (20,429 entries) concatenated with a reversed decoy database. Trypsin/P was specified as the cleavage enzyme, allowing up to one missed cleavage. Excision on the amino-terminal Met and carbamidomethylation on Cys were specified as fixed modifications. The false discovery rate (FDR) was adjusted to be less than 1%.
Missing value imputation
To address missing values in our dataset, we used the K-nearest neighbours imputation method using the KNNImputer function from the scikit-learn library in Python28.
Genetically engineered mouse models
The KrasLSL-G12D (ref. 29), Trp53flox (ref. 30), Trp53R172H (ref. 31), Pdx1-cre (ref. 32), Pdx1-Flp, KRasFSF-G12D, Trp53frt, Rosa26FSF-creER (ref. 14), Rosa26LSL-YFP (ref. 33), Rosa26mT/mG (ref. 15) and Grem1fl (ref. 34) mouse lines have been previously described. The VIM–GFP transgenic line developed by the Gene Expression Nervous System Atlas (GENSAT) Project was obtained from the Mutant Mouse Resource & Research Centers9. Spp1fl mice (NM-CKO-210205) were purchased from the Shanghai Model Organisms Center. These lines were intercrossed to generate the desired genotypes of both sexes on a C57BL/6 background in Charles River Laboratories. Mouse genotypes were determined using real-time PCR with specific probes designed for each gene (Transnetyx).
To activate CreERT recombinase after tumour onset, Pdx1-Flp;KrasFSF-G12D;Trp53frt/frt;Rosa26FSF-creER;Rosa26mT/mG;Spp1fl/fl (Spp1fl/fl KPFCT), Spp1WT/WT KPFCT, Grem1fl/fl KPFCT, Grem1fl/WT KPFCT, Spp1fl/flGrem1fl/WT KPFCT and Spp1fl/fGrem1fl/fl KPFCT mice were given tamoxifen through intraperitoneal injections at a dose of 100 mg kg−1 body weight, once a day for 5 days at 25 days of age. Pdx1–Flp;KrasFSF-G12D;Trp53WT/frt;Rosa26FSF-creER;Rosa26mT/mG;Spp1fl/fl (Spp1fl/fl KPhetFCT) and Spp1WT/WT KPhetFCT mice were given tamoxifen at the same doses at 100 days of age. Sample sizes were determined on the basis of the results of our preliminary experiments. Mice from autochthonous tumour models were selected according to their correct genotypes.
Mouse blood samples and ELISA
Plasma was isolated from the collected blood samples from mice after humane euthanasia. The samples were centrifugated at 12,000g for 10 min at 4 °C, and the supernatant was carefully transferred to a fresh centrifuge tube. Protein levels were quantified using a BCA kit according to the manufacturer’s protocol. ELISA was performed according to the Mouse & Rat Osteopontin (OPN) ELISA Kit–Quantikine protocol (R&D System). The plasma SPP1 concentration was calculated on the basis of the Mouse/Rat OPN standard.
Subcutaneous transplantation with mouse pancreatic cancer organoids
The immunocompromised Nu/Nu mice used have been previously described35, and adults (aged 8–12 weeks) obtained from Charles River Laboratories were used for transplantation.
Cells dissociated from organoids were subcutaneously injected in 50 μl cold growth-factor-reduced Matrigel (Corning). For Spp1 knockout organoid transplantation, a total 10,000 Spp1Δ/Δ KPF and Spp1WT/WT cells were injected into contralateral flanks of Nu/Nu mice. For Itgb3 knockout organoid transplantation, a total 100,000 Itgb3Δ/Δ KPF and Itgb3WT/WT cells were injected into contralateral flanks of Nu/Nu mice. For the inducible Grem1 deletion after Spp1 deletion analysis, we used CRISPR–Cas9-mediated knockout of Spp1 in GFP-labelled Grem1fl/fl KPFC organoids, and a total 100,000 cells dissociated from GFP-labelled Grem1fl/flSpp1Δ/Δ KPFC organoids were injected into the flanks of Nu/Nu mice. After tumours reached a size of 100 mm3, tamoxifen or peanut oil (vehicle) was intraperitoneally injected at a dose of 100 mg kg−1 body weight once a day for 5 days. For the inducible Grem1 overexpression after Spp1 overexpression analysis, we used Plv-SPP1–GFP and pINDUCER-GREM1 overexpression lentiviral vectors in KPFC organoids, and a total of 100,000 cells dissociated from Plv-SPP1–GFP and pINDUCER-GREM1 KPF organoids were injected into the flanks of Nu/Nu mice. After tumours reached a size of 100 mm3, doxycycline or saline (vehicle) was intraperitoneally injected at a dose of 2 mg kg–1 body weight twice weekly.
Tumour growth was monitored twice a week and mice were humanely euthanized when the tumour size reached a mean diameter of 1.2 cm. The tumours were weighed and fixed in 10% neutral buffered formalin (Cellstor) for histological analyses.
All animal experiments were approved by the Institute of Cancer Research, London’s Animal Welfare and Ethical Review Body and conformed to UK Home Office regulations under the Animals (Scientific Procedures) Act 1986, including Amendment Regulations 2012. The mice were housed in individually ventilated cages at constant temperature and humidity (23 ± 2 °C, 50–60%) in a pathogen-free controlled environment, with a standard 12 h light–12 h dark cycle and unlimited access water and food. For subcutaneous transplantation experiments, the maximum tumour size did not exceed the mean diameter of 1.2 cm, in compliance with project licence PP9490916.
Ultrasound-guided pancreas orthotopic transplantation
The injection technique was adapted from published literature36,37. Under anaesthesia, mice were placed in a supine position on a heated (37 °C) ultrasound platform. The platform was rotated 180° anticlockwise to position the injection mount towards the left upper quadrant of the abdomen. Ultrasound gel was applied to the upper part of the abdomen and the pancreas was located by using the left kidney and spleen as a reference. Once a clear image of the pancreas was located, a syringe was mounted into the holder and the needle was brought towards the field of view using the x axis micromanipulator. With an appropriate angle, the needle was pierced through the skin and pushed forward until the tip was in the pancreas. KPR172H/+C Spp1 overexpression (n = 10,000) cells and KPR172H/+C control (n = 10,000 cells) cells, suspended in 25 μl of 90% cold growth-factor-reduced Matrigel (Corning), were slowly injected into the pancreas using a 1 ml syringe with a 29 gauge needle. Successful injection was confirmed by the presence of hyperechogenic contrast from the Matrigel inside the pancreas. The needle was slowly withdrawn after 15 s of delay to prevent leakage.
Ultrasound imaging of pancreatic tumours
The precise acquisition of ultrasound images of pancreatic tumours was performed using a Microscan transducer MS-550D (VEVO 2100; Visualsonics). Mice were gently anaesthetized with 1.5–2.5% isoflurane in oxygen at a flow rate of 1 l min−1 and placed in a supine position on a preheated (37 °C) ultrasound platform. Pancreatic tumours were located and labelled using the 2D B-mode setting. For tumour volume measurement, consecutive 2D-mode images were acquired by scanning across the entire tumour at intervals of 0.2 mm using the 3D stage control system, and then reconstructed into a 3D volume. The heart rate, respiration rate and body temperature were monitored throughout the scanning process to ensure stable vital signs of the mice. The 3D tumour volume was calculated using the 3D analysis tool Vevo Lab (v.3.2.0).
Patient-derived organoid pancreatic cancer xenografts
Organoids were grown to 80–90% confluence before injection. Organoids were dissociated using TrypLE Express supplemented with Y-27632 (10 µM) into single cells and small clusters, and then counted using an automated cell counter. Cells were then resuspended to the indicated concentration in a 50% Matrigel and 50% organoid culturing medium. Next, 50 µl of the cell mixture was injected into the flank of male NSG mice aged 6 weeks. The tumour size was monitored twice weekly with callipers, and tumours were allowed to grow for about 2 months. Tumour volumes were calculated using the formula volume = length × width2/2, as previously described3. Mice were killed at either the study end point or when the tumour reached the maximum allowable volume (tumour volume = 1.5 cm2), and tissues were collected for downstream analyses. The animal use protocol listed was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC), Sun Yat-Sen University (approval no. SYSU-IACUC-2025-B0073).
SPP1 neutralization experiments
KPR172H/+CY mice (100 days old) were intraperitoneally injected 3 times a week for 3 months with mouse SPP1 antibody (BioxCell, clone MPIIIB10; 200 μg per mouse per injection) or control antibody (mouse IgG isotype control, Cell Sciences; 200 μg per mouse per injection) in independent experiments.
Mouse tumour cell isolation and organoid cultures
After humane euthanasia, mice were disinfected with 70% ethanol and pancreatic tumours were dissected and placed into ice-cold DMEM–F-12 containing 50 U ml–1 penicillin–streptomycin (Thermo Fisher Scientific). The tissue samples were minced on ice using sterile tissue scissors and then incubated in DMEM containing 1 mg ml–1 collagenase V (Sigma-Aldrich) in a shaking incubator for 1 h. After digestion, the process was halted by adding ice-cold G solution, and the cells were centrifuged at 300g for 5 min at 4 °C. Subsequently, the cells underwent further digestion with TrypLE (Thermo Fisher Scientific) for 10 min at 37 °C, followed by stopping the reaction with ice-cold 2% FCS in PBS and centrifugation. The cells were resuspended, filtered through a 40 μm nylon mesh, centrifuged again and then plated in Matrigel. Organoid medium was used as previously described38. To ensure the selection of cancer cells in the culture, for the first five passages, they were cultured in medium without EGF. After the fifth passage, complete organoid medium was used for culturing and experimentation.
Organoid staining
Organoids were grown in chamber slides (Ibidi) and washed twice with ice-cold PBS, fixed for 15 min in 4% paraformaldehyde in PBS, permeabilized in 0.5% Triton X-100 in PBS for 20 min and blocked in PBS containing 10% FCS (Gibco), 1% BSA (Sigma-Aldrich) and 0.2% Triton X-100 (Sigma-Aldrich). Primary antibodies were incubated in blocking buffer at 4 °C overnight. Fluorophore-conjugated secondary antibodies together with 3 μM DAPI were incubated in blocking buffer for 1 h at room temperature. The antibodies used are listed in Supplementary Table 2. Stained organoids were imaged on a Leica SP8 confocal microscope and analysed with Imaris (v.9.5.1).
Immunohistochemical staining
After standard dissection of mice, tumour tissue samples were collected and fixed in 10% neutral buffered formalin for 24 h. The tumours were then transferred to 70% ethanol for storage, followed by gradual dehydration in ethanol and embedding in paraffin. The paraffin-embedded tissues were sectioned into 4-μm-thick slices. H&E staining and DAB immunohistochemistry were conducted by the Histopathology Core Facility Breast Cancer Research at The Institute of Cancer Research using standard procedures. All staining was carried out using a Dako Autostainer Link48 (Agilent Technologies) immunostaining platform with epitope retrieval and simultaneous dewaxing using the Dako PT Link module (Agilent Technologies) according to the manufacturer’s instructions. Formalin-fixed paraffin-embedded sections were cut at 4 µm onto APEX adhesive slides (Leica Biosystems), dried at 60 °C for 60 min and subject to epitope retrieval at 97 °C for 20 min using either Dako Target Retrieval Solution pH 9 (Agilent Technologies, K800421-2) or pH 6 (Agilent Technologies, K800521-2) as described below. Primary antibodies were diluted in Dako FLEX antibody diluent (Agilent, K800621-2) and endogenous peroxidases were blocked using Dako REAL peroxidase block (Agilent, S202386-2). Antibody binding reactions were visualized using Dako DAB+ (Agilent, K346811-2) and sections were counterstained using Dako FLEX Haematoxylin (Agilent, K800821-2). The following antibodies were used for the indicated experiments: CK19 DSHB rat monoclonal TROMA III (primary dilution 1:200), epitope retrieval = pH 9, detection system = anti-rat N-histofine polymer-HRP (Nichirei Biosciences, 414311F); vimentin, Cell Signaling Technology rabbit monoclonal D21H3, 5741 (primary dilution 1:200), epitope retrieval = pH 9, detection system = Dako rabbit EnVision polymer-HRP (Agilent, K400311-2). Stained slides were imaged on a NanoZoomer (Hamamatsu Photonics) slide scanner and analysed with QuPath (v.0.4.2)39. Analyses and grading of PDACs were performed by a certified pathologist based on H&E staining. For liver and lung metastases, H&E-stained sections of paraffin-embedded tissues were analysed, and the number of metastases per section was quantified using ImageJ.
Immunofluorescence staining
Paraffin sections from tumour tissues were prepared as described above, and antigen retrieval was performed by heating in 10 mM sodium citrate (Sigma-Aldrich) buffer (adjust pH 6.0 by using hydrochloric acid). The sections were blocked using blocking buffer (PBS containing 10% FCS and 1% BSA from Sigma-Aldrich) at room temperature for 1 h, followed by overnight incubation with primary antibodies prepared in blocking buffer at 4 °C in a cold room. On the next day, fluorophore-conjugated secondary antibodies (1:200) along with 3 μM DAPI (Sigma-Aldrich) in blocking buffer were applied in the dark at room temperature for 1 h. A list of the antibodies used is provided in Supplementary Table 2. Before mounting, slides were incubated in 0.1% (w/v) Sudan Black B (Sigma-Aldrich) in fresh 70% ethanol to reduce background signal. Stained slides were imaged on a Leica confocal microscope or a NanoZoomer (Hamamatsu Photonics) slide scanner and analysed using Imaris.
CRISPR–Cas9-mediated knockout of target gene in organoids
The target gene was deleted in organoids using the CRISPR–Cas9 system. The protocol for engineering organoids was adapted and modified from a previous study40. Organoids were dissociated into single cells using TrypLE and diluted to 1 × 106 cells per electroporation. The cell pellet was then resuspended in a precise 100 μl solution containing BTXpress solution (BTX), 7.2 μg PB-CMV-MCS-EF1a-RFP-T2A-Puro (System Biosciences) or pPB[Exp]-CMV>Hygro (Vectorbuilder), 2.8 μg CMV-pHyPBase or pRP[Exp]-mCherry-CAG>hyPBase (Vectorbuilder), and 10 μg PX330 (provided by F. Zhang, Addgene, 4223062) with a target gRNA. The cell–DNA mixture was transferred to 2 mm electroporation cuvettes (Nepagene) and electroporated using a NEPA21 super electroporator (Nepagene), with settings described in our previous publication41. After electroporation, the cell suspension was transferred to complete medium containing Y-27632 (10 μM) without antibiotics. After 30 min, cells were centrifuged at 300g for 5 min at 4 °C, resuspended in Matrigel and cultured in antibiotic-free complete medium supplemented with 10 μM Y-27632. On day 5 after electroporation, organoids were selected with 2 μg ml–1 puromycin or 100 mg ml–1 hygromycin B. After 3 days of selection, organoids were flow sorted into single cells into a 96-well plate with 30 μl Matrigel per well. gDNA was extracted using a Purelink Genomic DNA Minikit per the manufacturer’s instructions (Thermo Fisher Scientific). PCR was performed using Q5 High-Fidelity 2× master mix (New England Biolabs) with pre-designed primers and gDNA. Subsequently, PCR products were amplified and cloned using a Zero Blunt PCR Cloning kit (Thermo Fisher Scientific) and sequenced by Sanger sequencing from Genewiz. A list of the gRNAs used is provided in Supplementary Table 3.
Flow cytometry
Organoids or tissues were dissociated into single cells as mentioned above. Single cells were suspended in FACS buffer (PBS containing 2% FCS). The cells were incubated with fluorophore-conjugated isotype control or experimental antibodies in FACS buffer on ice in the dark for 30 min. A list of the antibodies used is provided in Supplementary Table 2. Cells were washed 3 times with PBS, filtered through a 40 μm nylon mesh (Corning) and then incubated in FACS buffer containing 3 μM DAPI or propidium iodide. Single-colour and fluorescence minus one staining control were used as necessary. The cells were analysed using FACSymphony A5 (BD Biosciences) analyzer. Data were analysed using FlowJo 10 software. The gating strategy is shown in Supplementary Fig. 2.
RT–qPCR
RNA was extracted from organoids using either a RNAqueous-Micro Total RNA Isolation kit (Thermo Fisher Scientific) or a RNeasy Mini kit (Qiagen) following the manufacturer’s instructions. For BMP2 and LDN193189 treatment, organoids were starved in DMEM with 1% FCS overnight and treated with vehicle, 100 ng ml–1 recombinant BMP2 Protein (R&D Systems) or 100 nM LDN193189 (Tocris) for 12 h, followed by RNA extraction from the cells. cDNA was generated using an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. The cDNA was diluted 1:10 in distilled water, and 2 μl was used per qPCR reaction. PowerUp SYBR Green master mix (Thermo Fisher Scientific) was used for qPCR on an ABI7500 or QuantStudio 6 Flex (Applied Biosystems) system. Primers were designed using the Primer-BLAST tool at NCBI and made by Sigma-Aldrich. Housekeeping gene mRNA levels (Actb or Rplp0) were used for normalization. mRNA levels were quantified using the difference in Ct values and calculated using the \({2}^{-\Delta \Delta {C}_{{\rm{t}}}}\) method42. A list of the qPCR primers used is provided in Supplementary Table 4.
Western blotting
Matrigel was removed from cells on ice using Cell Recovery solution (Corning) following the manufacturer’s instructions. The Matrigel-depleted cells were lysed in ice-cold cell lysis buffer (NEB) as previously described3. After brief sonication, lysates were centrifuged at 13,000 r.p.m. for 15 min at 4 °C. Protein quantification was performed using a Protein Assay kit (Bio-Rad), and proteins were separated by SDS–PAGE on 10% gels (Bio-Rad), then transferred onto PVDF membranes (Bio-Rad) using a Trans-Blot Turbo system (Bio-Rad) according to the manufacturer’s instructions. The membranes were blocked in TBST containing 3% BSA at room temperature for 1 h, then incubated with primary antibodies prepared in TBST with 1% BSA overnight at 4 °C. HRP-conjugated secondary antibodies were subsequently incubated at room temperature for 1 h. Chemiluminescence was detected using Amersham ECL Prime Western Blotting Detection reagent (GE Healthcare) in an Amersham ImageQuant 800 imaging system. A list of the antibodies used is provided in Supplementary Table 2.
Lentiviral production and infection
The Itgb3 overexpression, Spp1 overexpression, SPP1 overexpression, control and GFP vectors were purchased from Vectorbuilder. GFP and inducible Grem1 were cloned into pLenti6 V5-DEST (Thermo Fisher Scientific) or pInducer20-Blast (Addgene, 109334) using the BP and LR Recombination System (Thermo Fisher Scientific). Lentiviral particles were produced in HEK293T cells using psPAX2 and pMD2.G helper plasmids (Addgene) and transfected into HEK293T cells using jetPRIME transfection reagent (jetPRIME). For organoid infection, a single-cell suspension was prepared and incubated with lentiviruses supplemented with 4 μg ml–1 polybrene (Santa Cruz Biotechnology) in a rocking incubator for 6 h. Cells were then seeded in Matrigel and antibiotic selection was applied 3 days later.
Chromatin immunoprecipitation
To predict transcription factor binding motifs in the Bmp2 promoter, DNA sequences ranging from −1000 to +500 bp from the transcription start site were scanned in the JASPAR 2000 database (http://jaspar.genereg.net/) using a relative profile score threshold of 80%. The JASPAR relative score is defined as 1 for the maximum-likelihood binding site21.
Organoids were dissociated and plated on 2D culture dishes and cultured in DMEM with 5% FCS. After exponential growth, the cells were switched to DMEM containing 1% FCS and starved overnight, followed by treatment with vehicle or 5Z-7-oxozeaenol for 6 h. The cells were fixed and crosslinked chromatin was prepared as previously described3. For immunoprecipitation, 10 μg chromatin was incubated with 10 μl anti-histone H3 rabbit IgG (CST, used as a positive control), 2 μl normal rabbit IgG (CST) or 5 μl of anti-RelA antibody (CST) overnight at 4 °C. Before immunoprecipitation, 2% of the chromatin (v/v) was used as input. Protein G magnetic beads were used to capture chromatin–protein–antibody complexes, followed by reversal of crosslinking to release chromatin and purification using a SimpleChIP Enzymatic Chromatin IP kit (CST). DNA was quantified by qPCR using primers targeting predicted Rela-binding regions at the Bmp2 promoter. The DNA level was normalized to the input, and the fold enrichment was calculated by normalizing to the control. A list of the chromatin immunoprecipitation qPCR primers is provided in Supplementary Table 5. A list of the antibodies used is provided in Supplementary Table 2.
RNA-seq analysis
For the RNA-seq analysis of Spp1 and Itgb3 WT and knockout organoids, RNA was extracted from organoids using a RNeasy Mini kit (Qiagen) following the manufacturer’s instructions. mRNA capture and library preparation were performed by Genewiz. Biological replicate libraries were sequenced on an Illumina NovaSeq platform at Genewiz, which generated an average of approximately 25 million 150 bp paired-end reads per sample. Sequence reads were processed using Trimmomatic (v.0.36) to remove possible adapter sequences and nucleotides with poor quality43. Reads were aligned to the mouse reference assembly to the Mus musculus GRCm38 reference genome available on ENSEMBL using STAR aligner (v.2.5.2b)44. Normalization of raw counts and regularized log-transformation were performed as previously described3.
Gene set enrichment analysis
For the RNA-seq dataset of Spp1 and Itgb3 WT and knockout organoids, GSEA (v.4.0.3) analysis was performed using the rlog-transformed counts, with the following settings applied: number of permutations = 1000, permutation type = gene set, enrichment statistics = weighted, metric for ranking genes = Signal2Noise. Normalized expression values of PDAC samples from the datasets GSE71729, ICGCarray, ICGCseq, TCGA, GSE62452 and GSE79668 were batch corrected using ComBat algorithm. Samples were grouped into SPP1high and SPP1low based on the median expression level. For GSEA, the following settings were applied: including the number of permutations = 1000, permutation type = phenotype, enrichment statistics = weighted, metric for ranking genes = Signal2Noise. The EMT gene set of 200 genes was checked for significant enrichment with an FDR q < 0.2.
scRNA-seq analysis
For human PDAC scRNA-seq data, fastq files of 24 PDAC and 11 normal pancreas samples were downloaded from the Chinese National Genomics Data Center (Genome Sequence Archive: CRA001160) and processed as previously described45. Uniform manifold approximation and projection plots were generated using the R package Seurat (v.3.2.2) with default settings. Cells were coloured according to cluster assignments from the reference. For evaluating SPP1 expression and its association with EMT in ductal cells (type 1 and 2), raw counts were normalized to counts per million (CPM) using the edgeR package in R (v.4.0.3) and log2[CPM + 1] transformed. Cells belonging to ductal clusters 1 and 2 were categorized into SPP1high and SPP1low based on expression levels.
Graphical abstracts
Diagrams in Fig. 1a and Extended Data Figs. 7a, 10a, 11a and 12 were created in BioRender (https://biorender.com).
Statistical analysis
All images and graphs represent the results of multiple experiments, which were conducted on different biological replicate samples and different dates. Each experiment was independently repeated. Differential gene expression analysis for RNA-seq data was performed using the edgeR package in R. All other statistical analyses were conducted using GraphPad Prism 8 for Windows. Specific statistical tests are specified in the figure legends. All t-tests tests, Fisher-exact tests and Mann–Whitney tests were two-sided.
Survival analysis
The survival plots for Spp1fl/fl KPFCT and Spp1WT/WT KPFCT mice receiving tamoxifen treatment and for Spp1fl/fl KPCY, Spp1WT/WT KPCY mice and SPP1 neutralization experiments were created and analysed using two-sided log-rank tests in GraphPad Prism 8. The following PDAC datasets were used: GSE71729, ICGCarray, ICGCseq, TCGA, GSE62452 and GSE79668. The normalized ComBat batch-corrected expression values were used for categorizing samples to SPP1high and SPP1low based on surv_cutpoint function in survminer R package. This function determines the optimal cut-off by maximally selected rank statistics.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.