Animal husbandry and ethics
All experiments using mice were approved and performed in compliance with the Swiss Law on Animal Protection (Loi fédérale sur la Protection des Animaux) under licence numbers GE45/20 and GE81/14. All animals were kept as a continuous backcross with C57BL6 × CBA F1 hybrids. The mice were housed at the University of Geneva Sciences III animal colony, with light cycles between 07:00 and 19:00 in the summer and 06:00 and 18:00 in winter. Temperatures were maintained between 22 °C and 23 °C, with humidity levels between 45% and 55%. The air was renewed 17 times per hour. Zebrafish (Danio rerio) were maintained according to standard conditions53 under a 14 h/10 h on/off light cycle at 26 °C, with set points of 7.5 and 600 μS for pH and conductivity, respectively. All zebrafish husbandry procedures were approved and accredited either by the Federal Food Safety and Veterinary Office of the canton of Vaud, Switzerland (no. VD-H23), by the animal committees of Rutgers University under protocol no. 201702646 or under the guidance of the Institutional Animal Care and Use Committee (IACUC) of Boston Children’s Hospital. AB, Tu and TL were used as wild-type strains and were obtained from the European Zebrafish Resource Center. The hoxdaDel(3DOM) and hoxdaDel(5DOM) mutants were generated for this study. Zebrafish embryos were derived from freely mating adults. Wild-type sibling hoxdaDel(3DOM) and hoxdaDel(5DOM) homozygous embryos were obtained by crossing the corresponding heterozygous mutant. Embryos were collected within 30 min after spawning and incubated at 28.5 °C in fish water, shifted to 20 °C after reaching 80% epiboly and grown at 28.5 °C to the proper developmental stage according to a previous study54. Pigmentation was prevented by treating the embryos with 0.002% N-phenylthiourea from 1 day post-fertilization (dpf) onwards. Sex was determined for animals used in the E18.5 UGS mouse experiments. Animals in other mouse experiments and in zebrafish experiments were not sexed. The sample size was not predetermined by a statistical test. Randomization and blinding were not conducted because the mutant and control animals were processed together in the same batch and grouped on the basis of their genotypes.
Generation of deletions in zebrafish
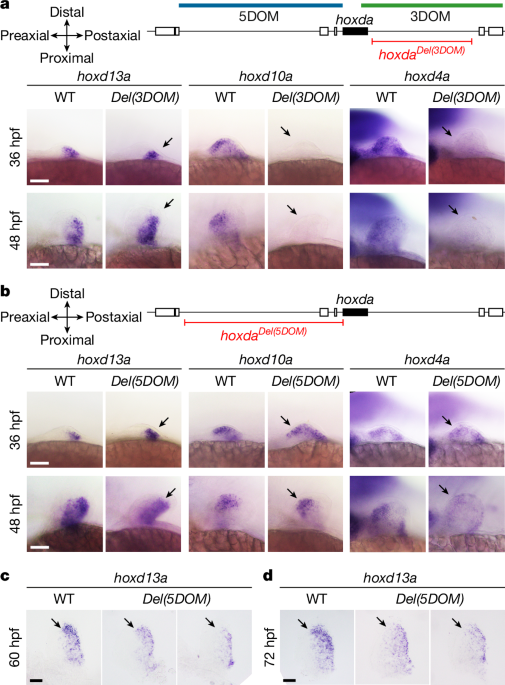
The hoxdaDel(3DOM) and hoxdaDel(5DOM) mutant alleles were generated using the CRISPR–Cas9 system described in a previous study55. The sequences of the CRISPR RNAs (crRNAs) used are listed in Supplementary Table 2. Loci were identified using the GRCz11 zebrafish genome assembly available on Ensembl. The corresponding genomic regions were amplified and sequenced from fin clips. Adults carrying verified target sequences were isolated and then selected for breeding to generate eggs for genome editing experiments. The guide RNA target sites were determined using the open-source software CHOPCHOP (http://chopchop.cbu.uib.no/index.php). Chemically synthesized Alt-R crRNAs and Alt-R trans-activating CRISPR RNAs (tracrRNAs) and the Alt-R Cas9 protein were obtained from Integrated DNA Technologies (IDT). To test the efficiency of these guide RNAs in generating the expected mutant alleles, we injected boluses ranging from 100 µm to 150 µm and containing 5 μM of the duplex crRNAs, tracrRNA and Cas9 ribonucleoprotein complex into the cytoplasm of one-cell-stage embryos. Injecting the ribonucleoprotein complex solution in a 100-µm bolus gave less than 5% mortality. With this condition, 30% of the embryos carried the 5DOM deletion and 15% carried the 3DOM deletion. For each condition, we extracted the genomic DNA of 20 individual larvae at 24 hpf for genotyping56. Identification of hoxdaDel(3DOM) and hoxdaDel(5DOM) mutants was performed using polymerase chain reaction (PCR). Amplification of evx2 was used as a control to confirm the presence or absence of 5DOM. The PCR mix was prepared using Phusion High-Fidelity DNA Polymerase (New England Biolabs), and primer sequences are listed in Supplementary Table 2. In parallel, 120 larvae per allele were raised to adulthood. To identify founders, F0 adults were outcrossed with wild type and 25 embryos were genotyped. Three and four independent founders were obtained for the hoxdaDel(5DOM) allele and hoxdaDel(3DOM), respectively. Two founders of each deletion were verified by Sanger sequencing (Supplementary Data 1) and used for further experiments.
Generation of knock-in reporter line
The endogenous hoxd13a reporter line (hoxd13aTg(hsp70:tdTomato)) was produced using a CRISPR–Cas9-mediated Gbait vector knock-in approach57,58. A guide targeting the coding region of exon 1 of hoxd13a (hoxd13a_KI_crRNA) was co-injected with a Gbait vector targeting guide (GFP_crRNA) and Gbait:hsp70l:tdTomato plasmid17. The injected embryos were screened for endogenous reporter RFP signal in expected hoxd13a expression domains, and positive individuals were raised to adulthood to outcross and recover F1 germline founders. To verify vector insertion and orientation in founders, genomic primers (hoxd13a_KI_F and hoxd13a_KI_R) were each paired with primers internal to the insert (LacZ_F and hsp70_R) for PCR and Sanger sequencing. The vector was oriented in the reverse direction relative to the endogenous promoter in the hoxd13aTg(hsp70:tdTomato) line, but reporter expression matched previously published in situ hybridization data and an hoxd13a knock-in line (hoxd13aegfp) generated independently by another research group59. Genotyping primers are listed in Supplementary Table 2.
Removal of CsB in cis to hoxd13a
Tg(hsp70:tdTomato)
To delete the CsB sequence from the chromosome carrying the hoxd13aTg(hsp70:tdTomato) endogenous reporter, each individual crRNA was duplexed with tracrRNA and injected at a final concentration of 6.25 μM with 1 μg Alt-R S.p. Cas9 Nuclease V3 (IDT). To estimate guide efficiency, DNA was extracted from four pools of three embryos each from 12 injected embryos and 12 control siblings and analysed using the T7 endonuclease 1 mismatch detection assay60. Embryos injected with efficient guides were raised to adulthood to outcross and identify founders. Guides flanking the CsB region (CsB_g1_crRNA and CsB_g2_crRNA) were injected into the hoxd13aTg(hsp70:tdTomato) background. The injected embryos were sorted by RFP signal at 1 dpf, and 16 positive animals from each clutch were screened for CsB removal using PCR with deletion-spanning primers (CsB_g1_F and CsB_g2_R) that did not amplify the intact locus under short elongation conditions. Clutches exhibiting a high frequency of CsB removal were raised to adulthood, and individuals were outcrossed to T5D wild type to obtain embryos carrying hoxd13aTg(hsp70:tdTomato)-Del(CsB) chromosomes. To identify CsB deletions in cis to the reporter, outcrossed embryos were sorted for RFP and then genotyped for the CsB deletion. One F0 injected parent (purple male 3) produced gametes with hoxd13aTg(hsp70:tdTomato)-Del(CsB) chromosomes at high frequency (approximately 25%), as well as gametes in which the CsB in cis to the reporter was left intact. Sanger sequencing of the deletion-spanning PCR product from 16 embryos revealed that each hoxd13aTg(hsp70:tdTomato)-Del(CsB) chromosome carried an identical deletion, suggesting clonality. Embryos resulting from outcrosses of this injected individual (purple male 3) were used in a subsequent expression analysis. The sequences of the crRNAs and genotyping primers used are listed in Supplementary Table 2. Sanger sequences of zebrafish founders are listed in Supplementary Data 1.
Quantification of hoxd13a
Tg(hsp70:tdTomato) expression
Outcrossed progeny with RFP signal from the endogenous reporter were collected at the 19-somite stage and at 72 hpf. Embryos were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (pH 7.4) for 2 h at room temperature with agitation in light-blocking containers, rinsed two times for 10 min each in PBS with 0.01% Tween 20 (PBST) and then incubated overnight at 4 °C in PBST with DAPI. The next day, the embryos were washed twice for 30 min each with PBST and processed for genotyping, as described above for the analysis of cloacal morphology of hox13 mutants, except that the head was removed for DNA extraction and the fins and trunk were retained for analysis. The embryos were imaged on a Zeiss LSM 800 confocal microscope to analyse hoxd13aTg(hsp70:tdTomato) expression. The laser and filter settings were optimized individually for each stage and tissue type to be compared, and then these settings were kept constant across CsB-intact and CsB-deleted individuals. For the 19-somite stage, the cloaca and tailbud were imaged simultaneously as a single piece of trunk, but for the 72-hpf animals, the fins, cloaca and tail were dissected and imaged separately. Maximum projection images were produced from each scan and then exported as TIF files for analysis in ImageJ61. Each image was cropped to a specific region of interest (ROI) containing the specific expression domain, and ImageJ was used to measure the mean grey value for pixels in the region. The ROI size for each tissue was as follows: 75 μm × 75 μm for cloaca (19-somite and 72 hpf), 250 μm × 250 μm for 19-somite tailbud, 200 μm × 200 μm for 72-hpf tails and 100 μm × 250 μm for 72-hpf pectoral fins. For each tissue, the average mean grey value was calculated from CsB-intact individuals and used to normalize signal intensity values so that the average CsB-intact intensity for each tissue was equal to 1. The average relative intensities for CsB-intact and CsB-deleted tissues were compared using Welch’s t-test in R (ref. 62).
Zebrafish hox13 mutant lines
Frameshift loss-of-function alleles hoxa13ach307, hoxa13bch308 and hoxd13a5bpins were previously generated11. The zebrafish lines were propagated and maintained, as described in a previous study63. To generate compound hox13 mutants, animals that were triple heterozygous for hoxa13a, hoxa13b and hoxd13a were intercrossed. The resulting larvae were fixed at 6 dpf in 4% PFA in PBS for 2 h at room temperature, with rocking agitation. After fixation, the larvae were rinsed twice for 5 min each in PBS with added 1% Triton X-100 (PBSX). To visualize the cloacal anatomy by labelling filamentous actin, the larvae were then incubated in PBSX with fluorophore-conjugated phalloidin (Sigma-Aldrich P1951; phalloidin-tetramethylrhodamine B isothiocyanate) added to a final concentration of 5 U ml−1 overnight at 4 °C, with rocking agitation. The larvae were then rinsed twice with PBSX for 1 h each.
For genotyping, the phalloidin-labelled larvae were cut in half, separating the head, yolk and pectoral fins from the cloaca and tail. The head half was used for genotyping, and the tail half was stored at 4 °C for later analysis. DNA was extracted from the head half by digesting tissue in proteinase K diluted to 1 mg ml−1 in 20 μl of 1× PCR buffer (10 mM Tris-HCl, 50 mM KCl and 1.5 mM MgCl2) for 1 h at 55 °C, followed by heat inactivation at 80 °C for 20 min. The digested tissue was then subjected to brief vortexing, and then 1 μl was used directly as template for genotyping PCR, with primers listed in Supplementary Table 2. For thermocycling, after an initial step at 94 °C for 2 min, reactions were cycled 40 times (15 s at 94 °C, 15 s at 58 °C and 20 s at 72 °C) and finished with 5 min at 72 °C. The PCR products were then heteroduplexed on a thermocycler by heating to 95 °C for 10 min and then gradually cooled by 1 °C every 10 s until a final temperature of 4 °C was reached. Heteroduplexed PCR amplicons were then run on a high-percentage agarose gel to determine the genotype by product size.
To analyse cloacal morphology, fixed phalloidin-labelled tails were imaged using a Zeiss LSM 800 confocal microscope. After acquiring a full confocal stack through the cloacal region, a midline frame that demonstrated the hindgut and pronephric duct morphology was selected. In a separate set of quantifications, juveniles were photographed using a Leica M205 FCA stereotype microscope, PLANAPO 1.0× zoom lens and Leica MC170 HD camera. Using the pencil tool in Illustrator, we traced the internal lumen of the hindgut and pronephric duct complex from the level of the proximal end of the median fin fold to the terminal exit. A perpendicular line was then drawn to measure the width of the complex. The lengths of these lines were measured using Illustrator and were used for statistical analysis.
Mutant mouse stocks
The following mouse lines used in this study were previously reported: Inv(Itga6-nsi)d11lac31, Inv(Itga6–attP) and tgBAC(HoxD)30, Del(HoxD)32 and Del(Atf2–SB1), Del(SB1–Rel5) and Del(Rel5–Rel1)2.
Whole-mount in situ hybridization
The zebrafish and mouse antisense probes used in this study are listed in Supplementary Data 2 and 3, respectively. For zebrafish, WISH was performed, as described56, at 58 °C for all riboprobes (hybridization temperature and saline–sodium citrate washes). Whole-mount embryos were photographed using a compound microscope (SZX10; Olympus) equipped with a Nomarski optics and a digital camera (DP22; Olympus). Genotyping of individual embryos was performed after photographic documentation using the primers listed in Supplementary Table 2. Wild-type and mutant embryos originated from the same clutch of eggs produced by heterozygote crosses and underwent WISH in the same well. Details on the number of embryos per experiment and genotype are provided in Supplementary Table 3. Murine urogenital systems were isolated from E18.5 embryos and processed following a previously reported WISH procedure64, with some specific adjustments. For proteinase K treatment, urogenital systems were incubated for 20 min in proteinase K diluted to 20 µg ml−1 in PBST. For the refixation step, a solution of 4% PFA containing 0.2% glutaraldehyde was used. The hybridization temperature was 69 °C, and the temperature of the post-hybridization washes was 65 °C. Staining was performed using BM-Purple (Roche; 11442074001) for approximately 4 h at room temperature.
Hybridization chain reaction
HCR in situ hybridization was performed, as previously described65, with slight modifications. Embryos were fixed in 4% PFA in PBS at 4 °C overnight with rocking, washed three times for 5 min in PBS with 0.1% Tween (PBST) and then dehydrated in methanol washes (25%, 50% and 75% in PBST) for 3 min each, followed by five 10-min washes and one 50-min wash in 100% methanol. The embryos were stored at −20 °C in methanol for at least 48 h before starting the hybridization protocol. The embryos were rehydrated in methanol (75%, 50% and 25% methanol in PBST), washed twice with PBST and pre-hybridized in hybridization buffer (Molecular Instruments) at 37 °C for at least 1 h. The embryos were then incubated in 200 μl of a hybridization solution with hoxd13a probes (IDT oPools; Supplementary Table 4) at a concentration of approximately 65 nM each overnight at 37 °C. After 18–24 h in probe solution, the embryos were washed four times for 15 min each using a probe wash buffer (Molecular Instruments) at 37 °C. The embryos were then washed twice for 5 min each at room temperature with 5× SCCT on a rocker before incubation in amplification buffer (Molecular Instruments) for at least 1 h. The amplification solution with B2 546 amplifiers (Molecular Instruments) was prepared by heating 3 μl of hairpin 1 (3 μM) and 3 μl of hairpin 2 (3 μM) to 95 °C for 90 s, followed by snap-cooling. After 30 min, hairpins 1 and 2 were mixed and added to 200 μl of amplification buffer. The embryos were incubated in amplification solution overnight at room temperature on a rocker. After 18–24 h of incubation in amplification solution, the embryos were washed at least four times for 30 min each with 5× SCCT at room temperature on a rocker. The embryos were stored at 4 °C in 5× SCCT for 1 day until they were genotyped and mounted for confocal microscopy. Before genotyping, the embryos were washed in PBST with DAPI for 1 h. DNA was extracted from the dissected head of each embryo. The pectoral fins were then microdissected using tungsten needles and mounted in PBST for confocal imaging using an inverted Zeiss LSM 800. Wild-type fins were used to optimize the laser and filter settings, which were maintained across all samples during data collection. After image acquisition, post-processing was performed on the maximum-intensity projections of each sample to reduce non-specific background signals. Specifically, the black value was changed from 0 to 50 uniformly for each image using the Zeiss Zen imaging software. These scans were then exported as TIF files for analysis in ImageJ61. The images were cropped to an ROI of 180 μm × 120 μm in size containing the hoxd13a fin expression domain. ImageJ was used to measure the mean grey value of ROI from each fin, and the average mean grey value was calculated from the wild-type fins. This average was used to normalize the signal intensity values such that wild-type fins had an average value of 1. The normalized relative intensities of wild-type and 5DOM deletion mutant fins were then compared using Welch’s t-test in R62.
Mouse genotyping
For extemporaneous genotyping, yolk sacs were collected and placed into 1.5-ml tubes containing rapid digestion buffer (10 mM EDTA (pH 8.0) and 0.1 mM NaOH) and then placed in a thermomixer at 95 °C for 10 min with shaking at 900 rpm. While the yolk sacs were incubating, the PCR master mix was prepared using Z-Taq (Takara; R006B) and primers (Supplementary Table 2) and aliquoted into PCR tubes. The tubes containing lysed yolk sacs were then placed on ice to cool briefly and quickly centrifuged at a high speed. The lysate (1 μl) was placed in the reaction tubes and cycled 32 times (2 s at 98 °C, 2 s at 55 °C and 15 s at 72 °C). The PCR reaction (20 μl) was loaded onto a 1.5% agarose gel, and electrophoresis was run at 120 V for 10 min. When samples could be kept for some time, a conventional genotyping protocol was applied using tail digestion buffer (10 mM Tris (pH 8.0), 25 mM EDTA (pH 8.0), 100 mM NaCl and 0.5% SDS) added to each yolk sac or tail clipping at 250 μl along with 4 μl of proteinase K at 20 mg ml−1 (Eurobio; GEXPRK01-15) and incubated overnight at 55 °C. The samples were incubated at 95 °C for 15 min to inactivate the proteinase K and stored at −20 °C until ready for genotyping. Genotyping primers (Supplementary Table 2) were combined with Taq polymerase (ProSpec; ENZ-308) in 25-μl reactions, cycled twice with Ta = 64 °C and then cycled 32 times with Ta = 62 °C.
Mouse RT–qPCR
UGSs were collected from E18.5 male embryos separately and placed in 1× diethyl pyrocarbonate–PBS on ice. A small portion of the remaining embryo was collected for genotyping. The UGSs were transferred into fresh 1× diethyl pyrocarbonate–PBS and then placed into RNAlater (Thermo Fisher Scientific; AM7020) for storage at −80 °C until processing. Batches of samples were processed in parallel to collect RNA using RNeasy extraction kits (QIAGEN; 74034). After isolating total RNA, first-strand complementary DNA (cDNA) was produced with SuperScript III VILO (Thermo Fisher Scientific; 11754-050) using approximately 500 ng of total RNA input. The cDNA was amplified with Promega GoTaq 2X SYBR Mix and quantified on a Bio-Rad CFX96 Real-Time System. Expression levels were determined by the difference between the cycle threshold (Ct) of the gene of interest (GOI) and the reference gene Tbp, calculated as dCt = Ct(GOI) − Ct(Tbp). They were normalized to 1 for each condition by subtracting each dCT from the mean dCT for each wild-type set. Finally, expression was evaluated by the power 2 minus this normalized dCT. Supplementary Table 2 contains the primer sequences used for quantification. RT–qPCR measurements were taken from distinct embryos. Box plots for expression changes and two-tailed unequal variance t-tests were produced in DataGraph 4.6.1. The boxes represent the IQR, with the lower and upper hinges denoting the first and third quartiles (25th and 75th percentiles). Whiskers extend from the hinges to the furthest data points within 1.5 times the IQR. The upper whisker reaches the largest value within this range, whereas the lower whisker extends to the smallest value within 1.5 times the IQR from the hinge.
Mouse RNA-seq
E18.5 male and female UGSs were collected by means of dissection separating the bladder from the UGS, including the proximal urethra in males and the vagina in females. Tissues were stored in RNAlater (Thermo Fisher Scientific; AM7020) and processed in parallel using RNeasy extraction kits (QIAGEN; 74034). RNA quality was assessed using an Agilent Bioanalyzer 2100 with RNA integrity number scores greater than 9.5. RNA sequencing libraries were prepared at the University of Geneva Genomics Platform using Illumina TruSeq Stranded Total RNA with Ribo-Zero Gold Ribo-deleted RNA kits to produce strand-specific 100-bp single-end reads on an Illumina HiSeq 2000. Raw RNA-seq reads were processed with Cutadapt v.4.1 (-a GATCGGAAGAGCACACGTCTGAACTCCAGTCAC -q 30 -m 15)66 to remove TruSeq adapters and bad-quality bases. Filtered reads were mapped to the mouse genome mm39 using STAR v.2.7.10a67 using ENCODE parameters with a custom gtf file68 on the basis of Ensembl version 108. This custom GTF file was obtained by removing readthrough transcripts and all non-coding transcripts from a protein-coding gene. Fragments per kilobase of transcript per million mapped read values were evaluated using Cufflinks v.2.2.1 (refs. 69,70) with the options –max-bundle-length 10000000 –multiread-correct –library-type ‘fr-firststrand’ -b mm10.fa –no-effective-length-correction -M MTmouse.gtf -G. Box plots depicting expression levels in distinct embryos were generated using the same methodology as that used for RT–qPCR.
ATAC-seq
Mouse and fish tissues were isolated and placed into 1× PBS containing 10% fetal calf serum on ice. Collagenase (Sigma-Aldrich; C9697) was added to 50 μg ml−1 and incubated at 37 °C for 20 min with shaking at 900 rpm. Cells were washed three times in 1× PBS. The number of cells was counted, and viability was confirmed to be greater than 90%. An input of 50,000 cells was processed according to a previous description36. Sequencing was performed on École Polytechnique Fédérale de Lausanne (EPFL) Gene Expression Core Facility (GECF) using an Illumina NextSeq 500. We analysed in a manner similar to a previous study71. Raw ATAC-seq paired-end reads were processed with Cutadapt v.4.1 (-a CTGTCTCTTATACACATCTCCGAGCCCACGAGAC -A CTGTCTCTTATACACATCTGACGCTGCCGACGA -q 30 -m 15)66 to remove Nextera adapters and bad-quality bases. Filtered reads were mapped on mm39 for mouse samples and danRer11 in which alternative contigs were removed for fish samples using Bowtie 2 v.2.4.5 (ref. 72) with the following parameters: –very-sensitive –no-unal –no-mixed –no-discordant –dovetail -X 1000. Only pairs mapping concordantly outside of mitochondria were kept (Samtools v.1.16.1) (ref. 73). The PCR duplicates were removed using Picard v.3.0.0 (http://broadinstitute.github.io/picard/index.html). The BAM files were converted to BED using bedtools v.2.30.0 (ref. 74). Peaks were called, and coverage was generated by MACS2 v.2.2.7.1 with –nomodel –keep-dup all –shift -100 –extsize 200 –call-summits -B. Coverages were normalized to million mapped reads.
ChIP–seq
Male UGSs were isolated and placed into 1× PBS containing 10% fetal calf serum on ice. ChIP–seq experiments were performed, as previously described75. Briefly, they were fixed for 10 min in 1% formaldehyde at room temperature, and the crosslinking reaction was quenched with glycine. Subsequently, nuclei were extracted, and chromatin was sheared using a water-bath sonicator (Covaris E220evolution ultrasonicator). Immunoprecipitation was performed using the following anti-H3K27ac (Abcam; ab4729) or anti-H3K27me3 (Merck Millipore; 07–449). Libraries were prepared using the TruSeq protocol and sequenced on an Illumina HiSeq 4000 (100-bp single-end reads) according to the manufacturer’s instructions. CTCF was reanalysed using datasets from previous studies43,71. The accession numbers are listed in Supplementary Table 5. Raw ChIP–seq single-end or paired-end reads were processed using Cutadapt v.4.1 (-a GATCGGAAGAGCACACGTCTGAACTCCAGTCAC for single-end reads and -a CTGTCTCTTATACACATCTCCGAGCCCACGAGAC -A CTGTCTCTTATACACATCTGACGCTGCCGACGA -q 30 -m 15)66 to remove TruSeq or Nextera adapters and bad-quality bases. Filtered reads were mapped on mm39 for mouse samples and danRer11 in which alternative contigs were removed for reanalysis of fish samples using Bowtie 2 v.2.4.5 (ref. 72) with the default parameters. Only alignments with a mapping quality above 30 were kept (Samtools v.1.16.1)73. Peaks were called, and coverage was generated by MACS2 v.2.2.7.1 with –call-summits -B (and –nomodel –extsize 200 for single-end reads). Coverages were normalized to million mapped reads/pairs.
Mouse enhancer–reporter assay
Transgenic embryos were generated, as described33. Primers were designed to amplify genomic DNA from the region around the observed ATAC and H3K27Ac peaks (Supplementary Table 5). These primers included extra restriction sites for either XhoI or SalI at the 5′ ends. The PCR fragments were cleaned using a QIAGEN Gel Extraction Kit (28704). The PCR fragment and the pSKlacZ reporter construct (GenBank X52326.1)75 were digested with XhoI or SalI and ligated together using the Promega 2X Rapid Ligation kit (C6711). Sanger sequencing confirmed that the correct sequences were inserted upstream of the promoter. Maxipreps of the plasmid were prepared and eluted in 1× IDTE (11-05-01-13). Pro-nuclear injections were performed, and embryos were collected at approximately E18.5 and stained for lacZ. UGSs were collected from E18.5 embryos in ice-cold 1× PBS in a 12-well plate. All steps were performed with gentle shaking on a rocker plate at room temperature. Tissues were fixed for 5 min at room temperature in freshly prepared 4% PFA. After fixing, the tissues were washed three times in 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet P-40 and 1× PBS for 20 min at room temperature. The wash solution was replaced with β-galactosidase staining solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2 hexahydrate, 0.01% sodium deoxycholate, 0.02% Nonidet P-40, 1 mg ml−1 of β-galactosidase and 1× PBS) for overnight incubation with the plate wrapped in aluminium foil to protect from light. The tissues were then washed three times in 1× PBS and fixed in 4% PFA for long-term storage. Images of embryos were collected using an Olympus DP74 camera mounted on an Olympus MVX10 microscope using Olympus cellSens Standard 2.1 software.
Mouse capture Hi-C sequencing
E18.5 male UGSs were collected, and collagenase-treated samples were crosslinked with 1% formaldehyde (Thermo Fisher Scientific; 28908) for 10 min at room temperature and stored at −80 °C until further processing, as previously described76. The SureSelectXT RNA probe design used for capturing DNA was performed using the SureDesign online tool by Agilent. Probes cover the region chr. 2: 72240000–76840000 (mm9) producing twice the coverage, with moderately stringent masking and balanced boosting. DNA fragments were sequenced on an Illumina HiSeq 4000 and processed with HiCUP v.0.9.2 on mm39 with –re1 ^GATC77, Bowtie 2 v.2.4.5 (ref. 72) and Samtools v.1.16.1 (ref. 73). The output BAM was converted to a pre-juicer medium format with hic2juicer from HiCUP. The pairs with both mates on chr. 2: 72233000–76832000 were selected, sorted and loaded into a 10-kb bin matrix with cooler v.0.8.11 (ref. 78). The final matrix was balanced with the option –cis-only. TADs were computed using HiCExplorer hicFindTADs v.3.7.2 (refs. 79,80) with –correctForMultipleTesting fdr –minDepth 120000 –maxDepth 240000 –step 240000 –minBoundaryDistance 250000. Data were plotted on mm39 (chr. 2: 73600000–75550000).
Zebrafish Hi-C sequencing
The HiC profiles were derived from a reanalysis of data from previous studies43,81. The accession numbers are listed in Supplementary Table 5. Reads were mapped on danRer11 in which alternative contigs were removed, and no selection of reads were performed. Valid pairs were loaded into a 10-kb bins matrix. TAD calling parameters were adapted to the smaller size of the genome: –chromosomes “chr9” –correctForMultipleTesting fdr –minDepth 35000 –maxDepth 70000 –step 70000 –minBoundaryDistance 50000. Data were plotted on danRer11 (chr. 9: 1650000–2400000) and on an inverted x axis.
CUT&RUN
Zebrafish samples were processed using a final concentration of 0.02% digitonin (Apollo; APOBID3301). Approximately 0.5 × 106 cells were incubated with 0.1 μg (100 μl)−1 of anti-H3K27ac antibody (Abcam; Ab4729) or 0.5 μg (100 μl)−1 of anti-H3K27me3 (Merck Millipore; 07-449) in digitonin wash buffer at 4 °C. The protein A–micrococcal nuclease was kindly provided by the Henikoff Lab (batch 6) and added at 0.5 μl (100 μl)−1 in digitonin wash buffer. Cells were digested in high-calcium buffer and released for 30 min at 37 °C. Sequencing libraries were prepared with KAPA HyperPrep reagents (07962347001) with 2.5 μl of adapters at 0.3 μM and ligated for 1 h at 20 °C. The DNA was amplified for 14 cycles. Post-amplified DNA was cleaned and size selected using 1:1 ratio of DNA:AMPure SPRI beads (A63881) followed by an extra 1:1 wash and size selection with HXB. HXB is equal parts 40% polyethylene glycol 8,000 (Thermo Fisher Scientific; FIBBP233) and 5 M NaCl. Sequencing was performed at EPFL GECF on an Illumina HiSeq 4000. Raw CUT&RUN paired-end reads were processed with Cutadapt v.4.1 (-a GATCGGAAGAGCACACGTCTGAACTCCAGTCAC -A GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT -q 30 -m 15) to remove TruSeq adapters and bad-quality bases66. Filtered reads were mapped on danRer11, in which alternative contigs were removed with Bowtie 2 v.2.4.5 (ref. 72) with the following parameters: –very-sensitive –no-unal –no-mixed –no-discordant –dovetail -X 1000. Only alignments with mapping quality above 30 were kept (Samtools v1.16.1) (ref. 73). PCR duplicates were removed by Picard v.3.0.0 (http://broadinstitute.github.io/picard/index.html). BAM files were converted to BED with bedtools v.2.30.0 (ref. 74). Peaks were called, and coverage was generated by MACS2 v.2.2.7.1 with –nomodel –keep-dup all –shift -100 –extsize 200 –call-summits -B. Coverages were normalized to million mapped reads.
Analyses of conserved sequences
Annotation of orthologous domains was performed using transcription start sites of orthologous genes, as reported in Supplementary Table 6. To identify conserved sequences between mouse and zebrafish, a pairwise alignment was done between the mouse genomic region chr. 2: 73600000–75550000 (mm39) and the zebrafish orthologous region chr. 9: 1650000–2400000 (danRer11) using discontinuous megablast. To reduce false positives, only reciprocal hits were considered. To display multispecies conservation levels, multiple alignment format files were generated between chr. 2 of the mouse genome (mm39) and contig chrUn_DS181389v1 of the platypus genome (ornAna2), chr. 7 of the chicken genome (galGal6), contig chrUn_GL343356 of the lizard genome (anoCar2), chr. 9 of the frog genome (xenTro10), contig JH127184 of the coelacanth genome (latCha1), chr. 9 of the zebrafish genome (danRer11), chr. 1 of the fugu genome (fr3) and the whole lamprey genome (petMar3). Details for the multiple alignment format generation are available on the GitHub repository (https://github.com/AurelieHintermann/HintermannBoltHawkinsEtAl2025; ref. 82). To facilitate visualization, a horizontal line was plotted for each species on each region.
Whole-genome alignments
Whole-genome alignments were performed using Progressive Cactus v.2.6.7 (ref. 83). The cactus command was used with default parameters to obtain the hierarchical alignment format. The hierarchical alignment was then projected on either zebrafish chr. 9 or mouse chr. 2 with cactus-hal2maf84 using –chunkSize 500000 and –noAncestor. The genome assemblies are listed in Supplementary Table 6.
Single-cell assay for transposase-accessible chromatin sequencing
The single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) bigwig files were downloaded from a previous study37 (Gene Expression Omnibus (GEO) GSE243256) where annotations were available. To annotate the cells from the cloaca, the raw matrix of single-cell RNA sequencing (scRNA-seq) from a previous study38 was downloaded from GEO (GSE223922) and stored in a Seurat object. Only 12,424 cells obtained at 14 hpf were kept. The data were normalized, and 3,000 variable features were extracted. Data were scaled. Uniform manifold approximation and projection (UMAP) and t-distributed stochastic neighbour embedding projections were calculated using the first 50 principal components. In parallel, scATAC-seq fragments of cells corresponding to 14 hpf were extracted from the general fragment file provided in a previous study38 on GEO (GSE243256). A new ArchR gene annotation was generated using the Lawson gtf v.4.3.2 (ref. 85) to match the scRNA-seq data from a previous study37, and the selected fragments were loaded into an ArchRProject with this genome. Iterative latent semantic indexing was computed with COR-Cut-off of 0.5. The clustering of scRNA-seq was then transferred to scATAC-seq using AddGeneIntegrationMatrix. The profile of the 38 cells whose transferred cluster corresponds to cloaca (endo.31) was generated with getGroupBW.
scRNA-seq
The matrix of the scRNA-seq atlas was downloaded from GEO (GSE223922; ref. 37) and the table with metadata. The matrix was loaded into a Seurat object using Seurat v.4.3.0 (ref. 86) in R v.4.3.0. Cells attributed to the ‘tissue.name’ ‘endoderm’ were selected. Normalization and principal component analysis were performed, as described in a previous study37. UMAP was performed on the top 70 principal component analyses and 50 nearest neighbours. UMAP coordinates and hox13 normalized expression of endoderm cells were exported to a file and plotted using ggplot2 v.3.4.4.
Software
The phylogenic tree was generated with http://timetree.org using the following species: Mus musculus, Protopterus, D. rerio, Carcharhinus leucas, Petromyzon marinus and Branchiostoma lanceolatum and subsequently edited using SeaView 4.7. Genomic tracks from next-generation sequencing were plotted using pyGenomeTracks 3.8 using custom gene annotations available at https://doi.org/10.5281/ZENODO.7510796 (ref. 68; mm39) and https://doi.org/10.5281/zenodo.10283273 (ref. 87; danRer11). RT–qPCR, RNA-seq and domain size quantifications were plotted in R using the ggplot package.
Ethical statement
All experiments involving mice were performed in agreement with the Swiss Law on Animal Protection (Loi sur la Protection des Animaux) under licence no. GE 81/14. For zebrafish, work was carried out either under a general licence of EPFL granted by the Service de la Consommation et des Affaires Vétérinaires of the Canton of Vaud, Switzerland (no. VD-H23) or was either agreed upon by the animal committees of Rutgers University under protocol no. 201702646 or under guidance of the Institutional Animal Care and Use Committee of Boston Children’s Hospital.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.