ASO design
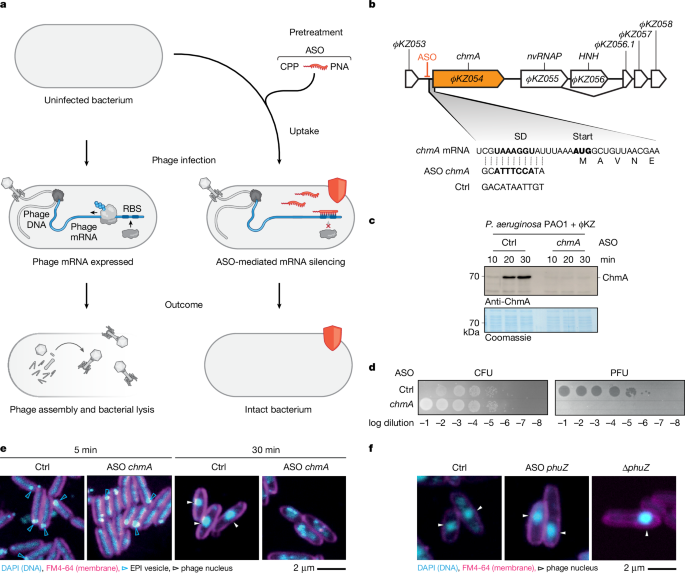
ASOs were designed to bind at the RBS using the MASON algorithm (https://mason.helmholtz-hiri.de)34 and the NCBI sequence and annotation files (ΦKZ: NC_004629.1, ΦPA3: NC_028999.1, PP7: NC_001628.1, SPO1: NC_011421.1, RAY: NC_041973.1, PA14: NC_008463.1, PAO1: NC_002516.2). ASO length was set to 11 nt and the allowed mismatches for off-targets were set to 4 nt. ASOs were selected based on the following scoring values: melting temperature (45–55 °C), low purine percentage (25–35%) and few predicted off-targets in distinct translation initiation regions of the phage (<3). The scoring values for the used ASOs are given in Supplementary Data 5. At least two ASOs were designed for each targeted gene. The control ASO sequence was GACATAATTGT (ctrl., JVPNA-79). ASOs were commercially ordered at Peps 4LS (Heidelberg) with a peptide backbone (PNA) and an N-terminal RXR (RXRRXRRXRRXRXB), KFF (KFFKFFKFFK) or TAT (GRKKRRQRRRYK) CPP. The initial concentration was adjusted to 1 mM in water based on the specific extinction coefficient using absorption. ASOs were stored at −20 °C. Prior to use, ASOs were thawed at room temperature, heated for 5 min at 55 °C and then cooled down to room temperature. All ASO sequences are listed in Supplementary Data 5, for example, JVPNA-72 for chmA knockdown, JVPNA-125 for nvRNAP knockdown, JVPNA-964 for PicA knockdown, and JVPNA-960 for PhuZ knockdown.
CFU and PFU assay
P. aeruginosa strains PAO1 (JVS-11761, DSMZ: DSM22644), PaLo8, PaLo9, PaLo39, PaLo44 (R. Lavigne laboratory, KU Leuven, Belgium), and PA14 (R. Lavigne laboratory, KU Leuven, Belgium) were grown in LB medium overnight at 37 °C and 220 rpm. Cells were inoculated 1:100 and grown in Mueller–Hinton medium at 37 °C and 220 rpm to an absorbance (optical density) of 0.3 (absorption given at 600 nm). ASOs were added at 6 µM final concentration to 50 µl cultures and incubated for 30 min. Cells were infected with ΦKZ at an MOI = 0.0001 and the cells were incubated for 180 min, which corresponds to three full replication rounds. Five microlitres of cell culture were diluted in series and spotted on LB plates and on 0.5% LB soft agar plates with the susceptible strain PAO1 (one volume of 0.5% LB soft agar at 42 °C, was mixed with 0.01 volume cells at an optical density of 0.5, and poured into a plate). Plates were imaged with the Typhoon 7000 phosphor imager (GE Healthcare) in fluorescence mode.
For jukA silencing in PA14, the cells were inoculated 1:100 in Mueller–Hinton medium, after 30 min cells were pretreated with 6 µM ASOs. The cells were infected after 150 min pre-incubation time with ΦKZ at an MOI = 0.0001, and incubated further for 180 min followed by CFU and PFU quantification.
For PP7 infection, PAO1 cells were inoculated 1:100 in Mueller–Hinton medium and treated at optical density 0.3 with 0.05 µM final concentration ASOs, additionally every 30 min 0.05 µM final concentration ASOs were added. Cells were infected 30 min after starting treatment with PP7 at an MOI = 0.00001, and cells were collected for CFU and PFU analysis after 120 min.
For RAY phage, P. agglomerans (DSMZ: DSM3493) cells were grown overnight in LB and were then inoculated 1:100 in Mueller–Hinton medium and grown at 37 °C, at optical density 0.3, cells were pretreated with 6 µM RXR–ASOs for 30 min. Cells were infected with phage at an MOI = 0.0001, followed by incubation for 300 min and spotting.
For SPO1 phage, B. subtilis 168 (BGSC: 1A1) was grown overnight in LB medium and then inoculated 1:100 in Mueller–Hinton medium, and grown at 37 °C, at optical density 0.3, cells were pretreated with 6 µM KFF–ASOs for 30 min. Cells were infected with phage at an MOI = 0.0001, followed by incubation for 180 min and spotting.
Immunoblotting
Infected cell cultures were mixed with final 1× SDS–PAGE loading dye (60 mM Tris/HCl pH 6.8, 0.2 g ml−1 SDS, 0.1 mg ml−1 bromophenol blue, 77 mg ml−1 DTT, 10% (v/v) glycerol) and were boiled for 10 min at 95 °C for denaturation. Protein samples were analysed by SDS–PAGE and blotted onto methanol-preactivated polyvinylidene (PVDF) membranes. As a loading control, we used Coomassie staining of a second gel where we loaded the same sample volume. ChmA was produced as previously described16 in E. coli BL21-CodonPlus (DE3)-RIL cells (Agilent Technologies, JVS-12280, chloramphenicol resistance) that were transformed with pET-M14(+) plasmid carrying the chmA gene with a N-terminal His-V5-TEV-tag (pMiG118). ChmA was purified as described before16. The tag could not be removed in the purification procedure. Commercial antibody sera were generated by Eurogentec. Rabbits were immunized with the purified ChmA protein. The rabbit serum (no. 2481) was used at a 1:10,000 dilution together with anti-rabbit HRP-conjugated antibody (Thermo Scientific, 31460) in a 1:10,000 dilution in 5% BSA/TBST for ChmA detection in immunoblotting. Antibody specificity was validated in immunoblotting by comparison between ΦKZ-infected and non-infected cells that yielded a defined band at 70 kDa corresponding to ChmA only in infected cells (Fig. 1c).
Microscopy
Agarose (0.85% (w/v)) was dissolved in fivefold water-diluted LB medium and boiled to melt. The liquid agarose was poured on microscope slides with one slide pair at each side as a spacer and one slide on top to form a closed gel slice as described61. After solidification, approximately 1 cm × 1 cm pads were cut. Cells were grown in Mueller–Hinton medium at 220 rpm and 37 °C to an A600 nm ≈ 0.3, then preincubated with 8 µM ASOs for 30 min, and infected with ΦKZ or ΦPA3 at an MOI = 10. Of note, ΦKZ ΔphuZ had a low titre owing to deletion of phuZ and could only be infected with an MOI of ~0.001. At indicated time points the phage replication cycle was quenched by cooling the cells on ice for 10 min and the cells were pelleted at 8,000g for 5 min. The supernatant was removed, and the cells were resuspended in 500 µl 4% paraformaldehyde and incubated for 15 min on ice. Afterwards cells were washed with PBS and were resuspended in 50 µl PBS for storage at 4 °C. For the imaging of the ΦKZ155 knockdown, we crosslinked bacteria in the medium with 2% glutaraldehyde for 30 min on ice, followed by the addition of 5% formaldehyde for 30 min on ice, as previously described62. Cells were stained with 16 µM FM4-64, and 360 nM DAPI and 5 µl were layered onto 1.2% agarose pads. The pad was placed with the side of application downwards into a µ-Slide 8 Well high Grid-500 (BD Biosciences). Transmission and fluorescence were detected with a confocal laser scanning microscope Leica SP5. Images were processed with ImageJ (1.53).
For the imaging of ΦKZ155–GFP, PAO1 cells were transformed with a plasmid (pLBu005) encoding ΦKZ155-gfp under the control of an arabinose-inducible pBAD promoter and selected on gentamycin plates. The cells (JVS-13713) were grown to an optical density of 0.25 and induced with arabinose at indicated concentrations followed by phage infection with an MOI = 10 at optical density 0.3. The collection, crosslinking and imaging of cells was conducted as described above for wild-type cells.
Proteomics
PAO1 cells were grown overnight in LB medium and were then inoculated 1:100 in Mueller–Hinton medium and grown at 37 °C and 220 rpm for 150 min to optical density 0.3. Cells were pretreated for 30 min with 6 µM ASOs against chmA, nvRNAP transcript (ΦKZ055), and picA. Subsequently, cells were infected with ΦKZ at an MOI = 5. At 2.5, 5.0, 7.5 and 10.0 min p.i. cells were collected with addition of 1/3 volume 4× Bolt SDS sample buffer (Invitrogen), and were immediately boiled at 95 °C for 5 min. Mass spectrometry sample preparation and measurement was conducted at the Proteomics Core Facility EMBL Heidelberg as previously described37. Proteins were quantified using label-free quantification (LFQ). Enrichment as log10-transformed P values was calculated with a two-sided Student’s t-test in MaxQuant Perseus (2.1.3)63.
RNA preparation and RNA-seq
PAO1 was grown overnight in LB at 37 °C 220 rpm. Cells were inoculated 1:100 in Mueller–Hinton medium at 37 °C and grown to an optical density of 0.3. 6 µM ASO (JVPNA-79 for control or JVPNA-72 for chmA inhibition) were added to 1.6 ml of cell culture and incubated for 30 min at 37 °C 220 rpm. Cells were infected with ΦKZ at an MOI = 5. At indicated time points, 250 µl were removed and incubated on ice. Infection efficiency was independently validated by confocal microscopy and CFU spotting with 50 µl of cells. RNA was isolated from 200 µl of cells using the RNAsnap procedure64. Two volumes of RNAprotect (Qiagen) were added and cells were incubated for 5 min. Cells were pelleted at full speed for 20 min at 4 °C and the supernatant was removed. The pellet was resuspended in 100 µl SNAP buffer (0.025% SDS, 18 mM EDTA, 1% β-mercaptoethanol, 95% formamide (RNA-grade)). Samples were incubated for 7 min at 95 °C, cell debris was pelleted at full speed for 5 min at room temperature, and the supernatant was transferred to a new tube. 1.5 volumes of ethanol were added to the supernatant and the sample was mixed by pipetting. The sample was loaded onto a miRNeasy mini column (Qiagen) two times and spun at full speed for 20 s at room temperature. Columns were washed two times with 500 µl RPE buffer (Qiagen) and spun at 8,000g for 1 min at room temperature. One final spin was used to dry the column in an empty tube. Thirty microlitres of RNase-free water was added to the column and the RNA eluted at 8,000g for 1 min at room temperature. The elution was repeated with the flow-through to recover more RNA. The RNA concentration was determined by absorption at 260 nm. RNA was stored at −80 °C.
RNA-seq was performed at the CoreUnit SysMed at the University of Würzburg. DNA was digested with DNaseI and the rRNA was depleted with the Lexogen RiboCOP META depletion kit. RNA library was prepared with the CORALL Total RNA-Seq Library Prep Kit V1 (Lexogen). The library was sequenced on a NextSeq2000 (Illumina) machine with a P1-seq kit (single-end 1× 100 bp, Illumina). RNA-seq analysis for the ChmA knockdown and the screen was conducted with READemption 0.4.3 and 2.0.4, respectively65. Reads were aligned for PAO1 and ΦKZ to NC_002516 and NC_004629, respectively. Enrichment of transcripts was calculated with the DeSeq2 module in READemption (2.0.4)65. Read coverage was illustrated with the Integrated Genomics Viewer (IGV)66.
We defined early and middle/late phage transcripts based on their significant enrichment (log2fold > 2 or log2fold < −2, −log10-adjusted P value > 10) between the control 35- and 10-min samples (Extended Data Fig. 6a, as in ref. 44).
Structure prediction
Structures were predicted from protein sequences using Google AlphaFold3 server (https://alphafoldserver.com/)67. This information is subject to AlphaFold Server Output Terms of Use found at https://alphafoldserver.com/output-terms (Google LLC). Ongoing use is subject to AlphaFold Server Output Terms of Use and of any modifications made. For the RNase HI domain structure in Fig. 4b, the predicted reference structure A0A2A2IBB4 was used from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/).
Complementation assay
PAO1 cells were transformed with pLBu019_ΦKZ155-TEV-3xFlag and pLBu021_ΦKZ155CDN-TEV-3xFlag and selected with gentamycin. Cells were grown overnight in LB medium with gentamycin. For the assay, no gentamycin was used in the medium. Cells were inoculated 1:100 in Mueller–Hinton medium. Cells were treated with 6 µM ASOs against control or ΦKZ155 for 30 min. For spotting, cells were infected with ΦKZ at an MOI = 0.0001, the complementation gene was induced with 0.2% arabinose, and cells were spotted at 180 min p.i. For imaging, cells were infected with ΦKZ at an MOI = 10, the complementation gene was induced with 0.2% arabinose, and cells were chemically crosslinked at 35 min p.i. and stained with FM4-64 and DAPI as described in the microscopy section and imaged.
In vitro translation
Template DNA was produced via PCR and Taq-polymerase followed by gel purification. Primers JVO-23244 and JVO-23245 were used to amplify wt ΦKZ155 with a T7 promoter from ΦKZ lysate, and for ΦKZ155D102N, pLBu021 was used as template. Template DNA (250 ng) was supplemented in 10 µl PURExpress in vitro protein synthesis kit mix (NEB, E6800). The reaction mix was incubated for 120 min at 37 °C and was subsequently used for assays.
Cleavage assays
RNA template (JVRNA-001, AUAUAAGGGAACAUAGAUAAACCCCUCCCUAAUAAAAUG) was labelled with 5′-32P. For the RNA–DNA duplex, the radiolabelled RNA was mixed with twice the amount of the reverse complement DNA (JVO-23273), boiled and slowly cooled down to room temperature in a water bath to anneal the duplexes. One picomole of RNA or RNA–DNA duplex was added to 2 µl PURExpress IVT mix that was translating for 2 h a control, ΦKZ155 or ΦKZ155(D102N). The mix was incubated for 1 h, mixed with 1 volume GLII buffer, boiled for 5 min, rapidly cooled down on ice, and loaded onto a 6% 6 M Urea–PAGE (19:1) gel that was run at 300 V for 2 h. The gel was transferred onto Whatman filter paper, vacuum dried, and a phosphor image screen was used to read out the autoradiogram on a Typhoon FLA7000 imager (GE Healthcare). As a positive control, we used a commercially available RNase H (NEB, M0297S).
Southern dot blotting
PAO1 cells were grown to an optical density of 0.3 in Mueller–Hinton medium and were pretreated with 6 µM ASOs against ΦKZ155 (ΦKZ) or gp176 (ΦPA3) for 30 min. Cells were infected at an MOI = 5 with ΦKZ. At indicated time points a fraction of the culture was removed and 1% SDS was added followed by boiling at 95 °C for 5 min. Subsequently, the DNA was extracted from the sample with phenol choloroform:isoamyl alcohol, and subsequently with one volume of chloroform. The aqueous phase was supplemented with 1.5 volume 1 M NaOH and 15 mM EDTA (pH 9). The sample was heated for 3 min at 95 °C and put on ice for 5 min. The solution was filtered with a dot blot apparatus through an equilibrated (0.3 M NaOH) and positively charged nylon membrane. Subsequently, the membrane was dried and the DNA crosslinked via exposition to UV light for 5 min. The membrane was equilibrated with hybridization solution two times, and a radiolabelled oligo was added for hybridization overnight starting at 60 °C for 1 h and then 48 °C overnight. The membrane was washed once for 15 min with 2× SSC and with 0.5× SSC, and a screen was used for phosphor imaging on a Typhoon FLA7000 imager (GE Healthcare).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.