Tissue procurement
All tissue was provided by the National Institutes of Health (NIH) NeuroBioBank and Banner Sun Health Research Institute Brain and Body Donation Program, which obtained written authorization and informed consent for all donors. Tissue collection and distribution for research purposes were done in accordance with protocols approved by the NIH NeuroBioBank (IRB protocol number: HM-HP-00042077) or the Human Brain and Spinal Fluid Resource Center (managed by the Sepulveda Research Corporation; IRB protocol number: PCC: 2015-060672, VA project number: 0002) and by the Banner Sun Health Research Institute Brain and Body Donation Program (WCG IRB protocol number 20120821). Tissue was collected from post-mortem, de-identified donors and thus this work is not considered by our Institutional Review Board to be research using human subjects. Cases were selected on the basis of RNA quality, age at time of death and absence of a history of neurological disease or evidence of neuropathology in the tissue. Brodmann area 9 or adjacent Brodmann area 46 of PFC was provided for each donor and used for both snRNA-seq and scWGS. To obtain the donor reference genomes, bulk DNA samples were collected from donor-matched tissues, which included heart, liver, muscle, cerebellum or cortex. Bulk DNA whole-genome sequencing (WGS) data for donors 1278, 4638, 1465, 4643, 5657 and 5817 (0.4-year-old male, 15-year-old female, 17-year-old male, 42-year-old female, 82-year-old male and 0.6-year-old male individuals, respectively) were obtained from previous studies8,58, along with bulk DNA WGS data for donor 5572 (70-year-old female individual)7.
Isolation of nuclei from fresh-frozen tissue samples
The nuclei isolation protocol was adapted from two previous publications59,60. All procedures were performed on ice or at 4 °C. Fresh-frozen samples were processed using a 7-ml glass Dounce homogenizer with approximately 20 mg tissue in 5 ml of filter-sterilized tissue lysis buffer (0.32 M sucrose, 5 mM CaCl2, 3 mM MgAc2, 0.1 mM EDTA, 10 mM Tris-HCl (pH 8), 0.1% Triton X-100 and 1 mM fresh DTT). The homogenized solution was loaded on top of a filter-sterilized sucrose cushion (1.8 M sucrose, 3 mM MgAc2, 10 mM Tris-HCl (pH 8) and 1 mM DTT) and spun in an ultracentrifuge in an SW28 rotor (13,300 rpm, 2 h, 4 °C) to separate nuclei.
For nuclei isolated for snRNA-seq, after spinning, the supernatant was removed and nuclei were resuspended (1% BSA in PBS plus 25 μl 40 U μl−1 RNAse inhibitor), then filtered through a 40-µm cell strainer. After filtration, nuclei were counted using trypan blue and an automated haemocytometer (Countess II; Invitrogen) and diluted to a concentration of 1,000 cells per µl.
For nuclei isolated for scWGS, the supernatant was removed and nuclear pellets were resuspended in ice-cold resuspension buffer (8.5 ml 1× PBS with 3 mM MgCl2 + 1 ml 1× PBS with 3 mM MgCl2 and 1% BSA + 500 µl sucrose cushion), filtered with a 40-µm cell strainer and then stained with an anti-NeuN antibody (directly conjugated to Alexa Fluor 488; Millipore MAB377X, clone A60; 1:1,250) and an anti-rabbit IgG Alexa Fluor 647 antibody as a negative control for 30 min. Using a BD Biosciences FACSAria Fusion machine and BD FACSDiva Software, forward scatter A (FSC-A) was first used to isolate large non-replicating cells. NeuN staining produced a bimodal signal distribution, distinguishing NeuN+ and NeuN− nuclei (Supplementary Fig. 13). Large neuronal nuclei, representing excitatory pyramidal neurons, were further identified by collecting the nuclei with the highest NeuN signal among the NeuN+ neuronal fraction, and gating for the population with the highest FSC-A signal and excluding Alexa-Fluor-647-high events7. This non-replicating high-FSC-A and high-NeuN population was confirmed to be an excitatory neuron population, comprising 2–5% of the total population of nuclei in each sample7.
Droplet-based snRNA-seq
Droplet-based libraries were generated using the Next GEM Single Cell 3′ v.3 or v.3.1 reagent kits (10x Genomics) and the Chromium Controller according to the manufacturer’s instructions. The resulting libraries were indexed with the KAPA Unique Dual-Indexed Adapter Kit (Roche KK8726) and sequenced on an Illumina NovaSeq 6000 with 150 paired-end reads by Genuity Science. Samples were prepared in batches of up to six donors at a time that always included male and female donors as well as mixed ages (Supplementary Table 3). To prevent age or gender bias in our batches, some samples have multiple biological replicates, prepared on different dates. A single replicate each from three distinct donors clustered abnormally during downstream analysis and was therefore excluded from analysis. After filtering, the only clusters exhibiting batch bias are those that are infant-specific and biologically driven (Supplementary Fig. 1). Because those cells were present only in infant donors, the only batches contributing to those clusters are those that included an infant.
In addition to data generated for this manuscript, we also included data that were previously published7: case 1465, a 17-year-old male individual. Single nuclei from the PFC were isolated by fluorescence-activated nuclear sorting using three gates (large NeuN+ nuclei, NeuN+ nuclei and DAPI+ nuclei) to generate three populations (large neurons, neurons and all nuclei). For each population, 16,000 nuclei were sorted into one well of a 96-well plate, which were then used to perform snRNA-seq using the Next GEM Single Cell 3′ GEM kit v.3.1 and the Chromium Controller (10x Genomics). The three resulting libraries were indexed using the 10x Genomics Dual Index Plate and sequenced on an Illumina NovaSeq S4. For our downstream differential expression analysis, all three populations were grouped together. Donor 1465 was excluded from analyses of cell-type proportion because the tissue had been subjected to fluorescence-activated cell sorting, which skewed the cell-type ratios.
scWGS of neurons using PTA
Single neuronal nuclei, prepared as described above, were whole-genome-amplified by PTA6,36 using the ResolveDNA Whole Genome Amplification kit (BioSkryb Genomics). First, nuclei were sorted into cold 96-well plates pre-loaded with 3 µl cold cell buffer (BioSkryb) one per well. Nuclei were lysed as per the kit protocol by the addition of 3 µl MS mix, followed by a brief spin-down, then 1 min of agitation at room temperature at 1,400 rpm on a plate mixer, then 10 min on ice. Next, 3 μl SN1 buffer was added to each well and the plate was again spun down and agitated at 1,400 rpm for 1 min. Next, 3 µl SDX buffer was added, and the plate was again spun and agitated at 1,400 rpm for 1 min. Then, the plate was incubated at room temperature for 10 min. Next, reaction mix and enzyme were added to each well, for a total reaction volume of 20 µl per well. PTA was performed for 10 h at 30 °C, followed by enzyme inactivation at 65 °C for 3 min. Amplified DNA was cleaned up using an in-house carboxyl magnetic bead clean-up solution (0.024 M PEG-8000, 1 M NaCl, 1 mM EDTA, 10 mM Tris-HCl pH 8, 0.055% Tween 20 and 1.5 ml Cytiva Sera-Mag SpeedBeads Carboxyl Magnetic Beads, hydrophobic per 50 ml). DNA yield was determined using the QuantiFluor dsDNA System (Promega). Samples were subjected to quality control by multiplex PCR for four genomic loci on different chromosomes as previously described8. Amplified genomes showing positive amplification for all four multiplex PCR loci were prepared for Illumina sequencing.
Libraries were prepared following a modified KAPA HyperPlus Library Preparation protocol described in the ResolveDNA EA Whole Genome Amplification protocol. In brief, the fragmentation step was skipped and end-repair and A-tailing were performed for 500 ng amplified DNA input. Adapter ligation was then performed using the SeqCap Adapter Kit (Roche, 07141548001). Ligated DNA was cleaned up using in-house beads and amplified through an on-bead PCR amplification step. Amplified libraries were selected for a size of 300–600 bp using double-size selection. Libraries were subjected to in-house quality control using a 5300 Fragment Analyzer Bioanalyzer for DNA fragment size distribution (Agilent Technologies). Successfully prepped samples were sent to Genuity Science for DNA sequencing, who further tested for quality using TapeStation (Agilent Technologies) before processing. Single-cell PTA-amplified genome libraries were sequenced on the Illumina NovaSeq 6000 platform (150 bp × 2) at minimum 20× coverage (Supplementary Table 12). scWGS of some neurons was performed at Harvard for previous publications6,7 (Supplementary Table 12).
Bulk DNA isolation
Genomic DNA was isolated using the QIAGEN DNA Mini kit (QIAGEN 51304) according to the manufacturer’s protocol for tissues. Approximately 25 mg of fresh-frozen tissue was minced on ice into small still-frozen pieces. Tissue was transferred to a dry-ice chilled sterile 1.5 ml microcentrifuge tubes with 180 μl of buffer ATL. Then, 20 ul of proteinase K (20 mg ml−1) was added before 4 h of agitation at 56 °C on a thermomixer (1,400 rpm). DNA isolation proceeded as written in the protocol with the inclusion of the optional RNase A treatment step. A small sample was sent for fragment analysing and gDNA quality assessment.
Bulk DNA library preparation and sequencing
Bulk DNA was isolated as described above and libraries were prepared following the KAPA HyperPlus library preparation protocol. The KAPA fragmentation step was included in the bulk processed gDNA samples. Bulk gDNA sample libraries were sent to Genuity Science and sequenced on the Illumina NovaSeq 6000 platform (150 bp × 2) at minimum 30× coverage and used as a reference genome against the case-match single-cell genomes. Bulk DNA for cases 1278, 4638, 4643, 5657 and 5817 was previously isolated and sequenced8 on an Illumina HiSeq X Ten platform by Macrogen Genomics or the New York Genome Center.
Analyses of snRNA-seq data
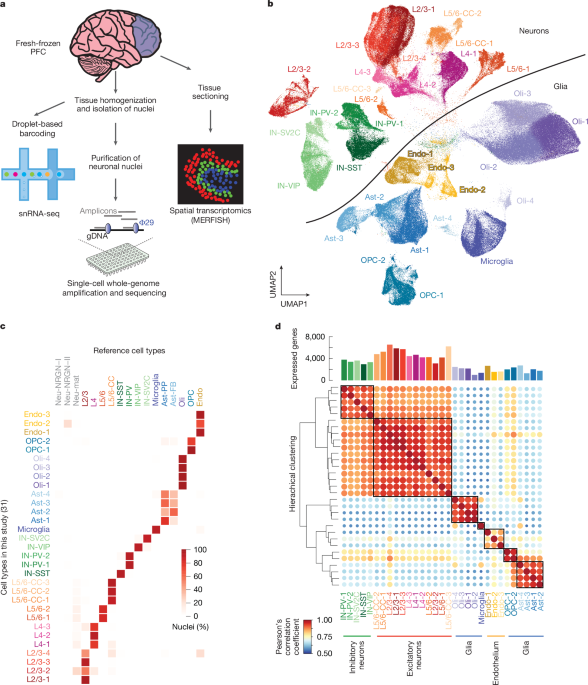
The snRNA-seq reads were aligned to the human genome and assigned to genes (GENCODE v.32) by Cell Ranger (v.6.0.2) with parameters –expect-cells=10000 –include-introns=true (ref. 61). The barcode and UMI solved counts were further processed with Seurat62 (v.4.3.0). The following filtering criteria were applied to each sample and cell: more than 100 cells in the sample; reads from mitochondrially encoded genes less than 5%; and more than 500 expressed genes in the cell. As discussed above, we further filtered samples ‘5817 200102’, ‘5288 200128’ and ‘5887 PFC 210601’ owing to their batch-driven, not cell-type-driven, clustering, removing them from downstream analysis. To minimize false discovery and focus on universal changes in ageing, mitochondrially encoded genes and genes in sex chromosomes were removed in the downstream analysis. The filtered data were log-normalized with a factor of 10,000. The top 8,000 variable features were selected for principal component analysis (PCA), clustering and uniform manifold approximation and projection (UMAP) analysis. The top 30 principal components and 0.5 resolution were used for k-nearest neighbours (KNN)-graph based clustering, yielding 39 clusters.
Each of the cells in this study was anchored to the cells from Velmeshev et al.19 using the RPCA method with the top 30 principal components19,62,63. For each of our 39 clusters, the percentages of cell types according to Velmeshev et al. were calculated, and the dominant cell types were used for each cluster. Those clusters with ambiguous cell types according to Velmeshev et al. were considered as artefacts and removed from the downstream analysis. We further defined marker genes for each cluster using the Seurat FindAllMarkers function by comparing each cluster with the remaining clusters, requiring expression in at least 25% of the cluster and a log2-transformed fold change greater than 0.25. For analyses in which excitatory neuron layer or inhibitory neuron subtype are not specified, layer- and subtype-specific clusters were combined and analysed as a group. Specifically, all neurons from the L2/3, L4, L5/6 and L5/6-CC clusters were combined into a non-layer-specific group of excitatory neurons, and neurons from the IN-SST, IN-SV2C, IN-PV and IN-VIP clusters were combined into a non-subtype-specific group of inhibitory neurons. Finally, we validated our cell-type assignment using the following marker genes (also shown in Supplementary Fig. 2): CUX2 for L2/3 neurons; RORB for L4 neurons; THEMIS for L5/6-CC neurons; TLE4 for L5/6 neurons; VIP, PVALB, SST and SV2C for inhibitory neuron subtypes; OLIG1 for oligodendrocytes; AQP4 for astrocytes; PDGFRA for OPCs; PTPRC for microglia; and CLDN5 for endothelia.
We identified changes in expression during ageing using the Seurat FindAllMarkers function. In brief, a Wilcoxon rank-sum test followed by multiple test adjustment was applied to determine significantly differentially expressed genes (q < 0.05) between adult and elderly donors for each cell type. We further filtered genes expressed in less than 25% of elderly cells and adult cells, or with a log2-transformed fold change less than 0.5. The same process was used to identify genes differentially expressed between infant cells and adult cells.
Continuous method to validate changes in expression during ageing
We used linear regression with sex as a covariate as an alternative method to determine continuous changes in expression during ageing. Average log-normalized expression levels and the age (in years) of each donor were used to build the linear model for each cell type. Genes with a slope less than −0.001 or greater than 0.001, a P value less than 0.05 and expressed in at least 25% of adult or elderly cells were considered as continuously changed genes during ageing. Both methods showed strong agreement on genes that go down during ageing across cell types, especially in excitatory neurons (Extended Data Fig. 5b and Supplementary Table 7). The linear model generally identified more genes that go up during ageing than the Wilcoxon test model, owing to the relatively strict log2-transformed fold-change cut-off of 0.5.
Analysis of transcriptome change during ageing using three groups
We investigated the transcriptome changes during ageing in a more continuous way, by dividing our non-infant donors into three groups: young adult (5 donors; under 40 years old); adult (6 donors; 40–69 years old); and elderly (6 donors; 70 years old or over). As shown in Extended Data Figs. 5a, 7a,e and 10a,b, the results generally matched our conclusions using the two-group comparison (elderly versus adult).
Transcriptional variability during ageing
Transcription variability is calculated by the coefficient of variation (CV). Specifically, for each gene in a specific cell type and a specific donor, the normalized expression levels (CPM) of all cells are used to calculate the CV, defined by the ratio of standard variation to the mean. The average CV of all genes is defined as the CV for a specific cell type within a particular donor. Comparing elderly and adult donors using a Wilcoxon rank-sum test showed a significant increase in transcriptional variability for IN-SST neurons but not for any other cell type.
Infant-specific analysis
To identify infant-specific changes in gene expression, we performed differential expression testing using the Seurat FindAllMarkers function as described above, comparing the infant-specific clusters (L2/3-2 and Ast-3) with the other non-infant-specific clusters of the respective cell type. The infant-specific upregulated genes, those with higher expression in the infant-specific cluster relative to the other clusters, were used for GO analysis (described below).
To determine changes in cell-type proportion, we used a Wilcoxon rank-sum test comparing the proportion of each cell type in infants to the remaining samples (adult and elderly). Donor 1465 (a 17-year-old male individual) was excluded from this analysis owing to the differences in nuclei preparation before snRNA-seq discussed above.
GO analysis
GO analysis of biological processes was performed on the differentially expressed genes for each cell type, both up and downregulated, using the R package gprofiler2 (v.0.2.3) with the correction method set to ‘fdr’ and source set to ‘GO:BP’ from the GO database. For each cell type, we used the active genes as the background gene set (indicated in the Supplementary Tables as control genes). Active genes were defined as those expressed in more than 25% of the cells to be consistent with the definition of a differentially expressed gene. Determination of the GO term categories shown in Figs. 2b and 3c was done manually (see Supplementary Tables 5 and 9 for mappings). To confirm the distinct GO enrichment profile in endothelial cells, we repeated the analysis after down-sampling. For each non-endothelial cell type, we chose the top 121 downregulated genes in elderly donors with the lowest FDR (121 matches the number of downregulated genes in the endothelial cells). There were fewer than 121 downregulated genes in oligodendrocytes, and thus down-sampling was not performed for this cell type. The GO down-sampling results are reported in Supplementary Table 10.
Random permutation test for shared downregulated genes in cell types from elderly donors
To test whether there are significantly more genes downregulated in at least one excitatory neuron, at least one inhibitory neuron and at least two glial cell types than expected, we performed a random permutation test. We randomly picked the same number of expressed genes to designate as downregulated for each cell type, using a minimum expression cut-off of 25% of the adult cells and 20% of the elderly cells, and recorded the number of shared genes as the expected value. A total of 1,000 permutations were performed, and all of the tests yielded fewer shared genes than observed in our data, generating a P value of less than 0.001.
Identification of sSNVs in neurons
To identify sSNVs, we used both scWGS and corresponding bulk WGS data. scWGS and bulk WGS data were first processed accordingly to the GATK (v.4.1.8.1) best practices64. In brief, reads were aligned to the human genome using bwa-mem (v.0.7.12) with default parameters. PCR duplicates were then filtered using Picard, and the remaining reads were recalibrated with GATK BaseRecalibrator and ApplyBQSR. Genotypes were then identified with GATK HaplotypeCaller and GenotypeGVCFs. Finally, sSNVs were identified by comparing the scWGS data with corresponding WGS data from bulk tissues using SCAN2 with the following parameters: –snv-min-sc-dp 5 –snv-min-bulk-dp 106. Common SNPs from dbSNP (v.20180418) and phasing information from the 1000 Genomes Project (v.3) were used as a reference panel while running the SCAN2 pipeline. We estimated the FDR for SCAN2 as 8.6% in a previous publication6.
Signature analysis of sSNVs
We performed signature analysis for sSNVs using the R package MutationalPatterns (v.3.16.0)65. We first calculated the spectrum of sSNVs in the 96-trinucleotide contexts for each neuron from all donors. A non-negative matrix factorization (NMF) was applied to the spectrum of sSNVs and the signatures were identified. After applying various numbers of signatures in the practice, ranging from one to eight, we found that two signatures yielded the best performance with regard to stability and reconstruction errors (Supplementary Fig. 12). The signatures (A1 and A2) were then compared with the COSMIC v.3 signatures, and cosine similarities between signatures were calculated. To confirm the reproducibility of our signature analysis, a second method, SignatureAnalyzer, was used with default parameters. SignatureAnalyzer identified similar signatures to those identified by MutationalPatterns.
Enrichment and strand bias of sSNVs in genic features and chromatin states
To calculate the enrichment of sSNVs in genes and intergenic regions, we first simulated random controls with the same mutation spectrum as sSNVs restricted to suitable regions (that is, with enough depth) in our scWGS and bulk WGS dataset. The numbers of sSNVs and random controls at genes and intergenic regions were then calculated. NMF, using the R package MutationalPatterns, was further applied to sSNVs and random controls at genes and intergenic regions to trace the contribution of signatures A1 and A2. Genes were divided into five groups according to their transcriptional activity (CPM) in neurons and glia cells from our snRNA-seq data. The same enrichment analysis was also done over the 15 chromatin states in the human dorsolateral PFC from Roadmap66. To test for strand bias in sSNVs, we used the UCSC table browser to identify all RefGene transcripts associated with single-neuron sSNVs. Only sSNVs that had known transcripts all going in the same direction were considered. Transcriptional directions for sSNVs that overlapped a transcript were tallied, and the numbers collapsed to report only one complement of each base pair (T>A, T>C, T>G, C>A, C>T and C>G).
DepMap analyses of the effects of upregulated and downregulated genes on cell viability
The requirement of each gene in overall cell viability was determined using the Cancer Dependency Map (DepMap; version Public 22Q4), which provides the cell viability effect of each gene knockout across 1,078 cancer cell lines of varying origin67. Specifically, cell viability is determined by performing whole-genome pooled CRISPR screening across each cell line, and on the basis of the fold change in the abundance of cells containing Cas9 and guides against each specific gene. For example, if cells transduced with Cas9 and guides against a particular gene were depleted after the screen, this would indicate an essential gene. The overall effect of gene knockout for a given cell line is quantified using a cell population dynamics model called Chronos68, which incorporates the efficacy of each guide and copy number correction (CRISPR toxicity unrelated to gene function can occur when high copy numbers are subjected to CRISPR-mediated strand breaks) to provide an overall ‘gene effect score’ that indicates the probability that a given cell line is dependent on the gene for survival69. Notably, a value of −1.0 corresponds to the median gene effect score of all common essential genes, whereas a cell line is considered dependent if the gene effect score is ≤ −0.5. Positive values would indicate increased cell viability or proliferation after loss of the gene.
Among the upregulated and downregulated ‘hits’ from the snRNA-seq, those encoding long non-coding RNAs, non-coding RNAs or pseudogenes are not covered in the DepMap essentiality analyses and thus were not analysed for effects on gene viability. Likewise, several coding genes (CECR, NEFL, FTH1, COX4I1, SH3RF3, BMP2K, SHISA8, MYRFL and RPS3A) did not have CRISPR screen data yet available, and were not analysed.
Defining housekeeping and neuron-specific genes
We first calculated the average logged CPMs for each gene in excitatory neurons, inhibitory neurons, microglia and endothelia. Then we defined housekeeping genes as genes with a difference of less than 0.1 between the four cell types that also had an average logged CPM greater than 0.1 in each cell type. The genes that fit these criteria also have an average logged CPM greater than 0.1 in oligodendrocytes, OPCs and astrocytes. The neuron-specific genes were defined as those genes with average logged CPMs higher than 0.2 in both neuron groups and lower than 0.1 in microglia and endothelia.
Determining what drives transcriptome change during ageing
To determine which feature is likely to drive expression change during ageing, we constructed a multiple linear regression model to estimate the contribution of genetic and transcriptomic features to the expression change during ageing. To avoid the effect of non-expressed genes, we only assessed genes whose average logged CPM is at least 0.1 in excitatory neurons, inhibitory neurons, microglia, endothelia, oligodendrocytes, OPCs and astrocytes. Gene length, exon length, expression in each cell type and expression specificity for each cell type and neurons were used to build the regression model to predict the fold change of gene expression between elderly and adult. Expression specificity was calculated by the normalized expression in each cell type divided by the average normalized expression in the remaining cell types. Expression specificity for neurons was calculated by comparing the average expression in neurons and the average expression in glia cells. We also included broad cell specificity in the model, defined by the sum of the difference between maximum expressed cell type and other cell types, divided by (number of cell types used – 1). As the number of reads captured for each gene could be biased towards gene or exon length, we also included gene and exon length-corrected normalized expression levels in each cell types as input features. Bulk sequencing is also a quantitative way to measure absolute expression levels. Thus, we included the expression levels (TPM; transcripts per million mapped reads) in human frontal cortex from the GTEx portal70. The squared correlation coefficient between the model prediction and observed fold change of expression, an indicator of model performance, ranged from 0.23 in microglia to 0.54 in excitatory neurons. Among all features assessed, gene length yielded the highest correlation coefficient, suggesting that it has a key role in determining expression change during ageing in neurons and glia cells.
Validation of snRNA-seq results using published data
No other single published study on human PFC spans the same age range as ours, so we looked to two different datasets for validation of our results. Herring et al.25 includes PFC from 22 gestational weeks to 40 years old. To validate our infant-specific clusters, we obtained raw snRNA-seq reads from the Herring paper (publicly available at GSE168408) and processed them using Cell Ranger (v.7.0.1) with the following parameters: “–include-introns true –nosecondary”. We filtered and clustered the data in the same way as we did with our own (described above), using Velmeshev et al. as our reference for cell-type identification, confirming the presence of infant-specific astrocytes and excitatory neurons in a larger sample size. We compared the expression of the infant-specific differentially expressed genes from our own data (methods described above) with Herring data for infants (prenatal samples to 2 years) and adults (15–40 years), validating our findings of infant-specific clusters and their respective gene-expression profiles.
To validate the changes we described in the elderly brain, we used control PFC (BA46) data from Ling et al.29, which includes donors aged 22–97 years. We downloaded their publicly available raw counts matrix for each cell type from NeMo (https://assets.nemoarchive.org/dat-bmx7s1t) and normalized the expression levels using the same strategy: to total number of reads for each cell with a factor of 10,000. We then compared the expression of genes of interest from our data in elderly and adult brains in the Ling data. Specifically, we assessed whether common genes are downregulated in elderly cells, and whether the decrease of expression during ageing is associated with gene length.
We also used the control PFC data from Mathys et al.30 to validate our findings. We downloaded their publicly available raw counts matrix for each cell type from the Alzheimer’s disease and ageing brain atlas data repository (https://compbio.mit.edu/ad_aging_brain) and normalized to the number of UMI reads per cell per 10,000 UMI reads. This dataset comprises 189 individuals, and includes only elderly donors (over 70 years old). We then compared the expression of downregulated genes, common genes and short and long genes in our adult donors, our elderly donors and the elderly donors from Mathys et al. The results were consistent with our own dataset: common genes and short genes showed decreased expression in neuron and glia cells from elderly donors. We also compared the expression levels of genes in donors aged 70–79 years and 80 years and over from Mathys et al., and did not find a significant change.
MERFISH: sample preparation and imaging
Spatial transcriptomics was performed in two batches using two versions of the MERFISH platform. Data from each batch were analysed separately and not integrated into a single analysis. For batch 1, three adult donors and three elderly donors were selected for spatial transcriptomics on the basis of RNA integrity number, tissue availability and sex. Vizgen’s protocol for the sample preparation was followed with the following modifications. Brains were sectioned and mounted on Vizgen MERSCOPE slides. After adhering to the coverslip, samples were fixed in prewarmed 4% paraformaldehyde in 1× PBS for 30 min at 47 °C, followed by three washes in 1× PBS for 5 min each at room temperature. Samples were dried for one hour at room temperature. The samples were then incubated overnight in 70% ethanol at 4 °C to permeabilize the tissue. Samples were photobleached for 6 h at room temperature in the Vizgen Photobleacher. Next, the Vizgen sample preparation protocol for FFPE tissues was followed, beginning with anchoring pretreatment (step 3 in Vizgen protocol version 9160012 Rev D). After RNA anchoring, the tissue was embedded in gel embedding solution (containing 0.5% ammonium persulfate, 0.05% TEMED and Vizgen’s gel embedding premix) and incubated for 22 h with tissue clearing solution (Vizgen Clearing Premix and 1:100 proteinase K) at 47 °C. The probe library was applied to the sample and incubated for 48 h at 37 °C. Finally, the samples were washed, incubated with DAPI and polyT solution for 15 min at room temperature and washed with formamide wash buffer for 10 min at room temperature. For the imaging, the MERSCOPE 500 gene imaging kit was activated with 250 μl imaging buffer activator and 100 μl RNAse inhibitor. Fifteen millilitres of mineral oil was added through the activation port, the instrument was primed and the imaging chamber was assembled according to the MERSCOPE user guide. A 10× low-resolution DAPI mosaic of the sample was acquired, and the imaging area was selected for data acquisition.
For MERFISH batch 2, an infant and three adult donors were selected on the basis of RNA integrity number, tissue availability and sex. All tissue processing steps were performed as described above, but imaging was performed on a MERSCOPE Ultra instrument. Owing to uncertainty in the back compatibility between instruments, these four samples were treated as their own set of data and never compared with the six-sample cohort processed on the older instrument.
MERFISH: post-imaging data processing and analysis
For batch 1, after the MERSCOPE run, the data were decoded using Vizgen’s analysis pipeline integrated within the MERSCOPE system. The Vizgen post-processing tool (VPT, Vizgen) was used to improve cell segmentation with a combination of pre-filtering with a Gaussian filter and the CellPose algorithm. For batch 2, the four samples imaged on the MERSCOPE Ultra were not subjected to additional processing using the VPT, because cell segmentation using CellPose was performed by the MERSCOPE Ultra instrument.
After cell segmentation, only cells with volumes greater than 200 μm3 were retained for downstream analysis. The cell × gene count matrix was then analysed with the Seurat R package (v.5.1.0) for cell-type assignment. Two datasets (one focusing on elderly versus adult, and one on infant versus adult) were analysed separated using the same pipeline. Specifically, PCA was performed using the count matrix, after filtering cells with fewer than 100 transcript counts, followed by logCPM transformation. We then performed UMAP and KNN clustering analysis using the top 30 principal components. The resolution of KNN clustering was set to 0.3, yielding 15 clusters in batch 1 and 16 clusters in batch 2. Each cluster was then assigned a specific cell type according to the expression of marker genes (Extended Data Fig. 6b and Supplementary Table 11).
In batch 1, clusters 12 and 14 showed mixed expression of marker genes, preventing cell-type assignment, and were removed from downstream analysis. Cluster 1 was composed of excitatory neurons that could not be assigned to a specific layer owing to mixed expression of layer-specific markers. To investigate the transcriptome change during ageing, we compared gene expression between adult and elderly donors for each cell type. We normalized gene expression first to cell volume (molecules per 2,000 μm3) and then to the average expression of a set of control genes that are stably expressed during ageing (Supplementary Table 11). The control genes were defined as genes with a log2-transformed fold change of expression between elderly and adult donors >−0.3 and <0.3 in our snRNA-seq dataset.
In batch 2, clusters 9, 10, 12 and 15 showed mixed expression of marker genes and were removed from downstream analysis. Different layers of excitatory neurons and different subtypes of inhibitory neurons were analysed together because many of them could not be assigned to a specific layer owing to mixed expression of layer-specific markers. We investigated transcriptome change during brain development using the same strategy as we did in the elderly and adult dataset. Gene expression between infant and adult donors was compared, after normalizing to cell volume and a set of control genes stably expressed in infant and adult donors.
MERFISH gene panel selection
The gene panel used for MERFISH was composed to validate initial snRNA-seq findings generated from 13 donors, focusing on differences in the elderly and adult donors. It is composed of 70 marker genes (used to identify cell types), 33 short housekeeping genes, 33 long housekeeping genes, 24 short neuron-specific genes, 21 long neuron-specific genes, 9 ribosomal-protein genes, 10 nuclear-encoded mitochondrial genes, 11 DNA damage repair genes and 35 other genes of interest (Supplementary Table 11). All short housekeeping and neuron-specific genes came from the first length decile of their respective gene groups and all long housekeeping and neuron-specific genes came from the tenth length decile of their respective gene groups. After the addition of six donors to our snRNA-seq data, our housekeeping and neuron-specific gene lists changed slightly, although the method used to generate the list did not, and not all of the neuron-specific and housekeeping genes in the MERFISH panel met the criteria. The MERFISH gene tab of Supplementary Table 11 reports the decile according to the original housekeeping and neuron-specific lists used to generate the panel. If the gene is present on the current list, the corresponding decile is reported in parentheses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.