Mice
Mice were bred and housed in the local animal facilities of the Centre Régional d’Exploration Fonctionnelle et de Ressources Expérimentales (CREFRE) in pathogen-free sanitary conditions, with free access to water and food, and were submitted to alternating cycles of 12 h of light and darkness at an ambient temperature of 22 °C. C57BL6j mice were purchased from Janvier Labs or Charles River laboratories. Mcpt5–cre66, iDTA, AdKOv2 and Cdh5–creERT2;mTmG mice were kindly provided by A. Roers (Heidelberg University Hospital), J.-C. Guéry, P. Val and F. Lenfant, respectively. On the basis of the results obtained in our laboratory in models of atopic dermatitis9, we estimate that, on the basis of the difference in the mean values of the clinical scores and with a predicted 3.5 standard deviation for individual measurements, we need six mice per group to give us 90% power to detect a difference in clinical score at statistical significance of *P ≤ 0.05 using a two-tailed unpaired t-test. Sample sizes for the behavioural tests were chosen to ensure equal numbers of male and female animals and equal group sizes, consistent with previous publications67. Sample sizes for the in vitro culture experiments (DRG neuron cultures and mast cell activation assays) were also selected to ensure equal numbers of replicates per experimental condition. The number of biological replicates was on the basis of previous publications using similar methods9,23, providing sufficient power to detect meaningful effects while accounting for biological variability. To minimize bias, after allocating CT and PS groups (to ensure equal representation of groups), the investigators were blinded to the group and/or genotype of the mice whenever possible during animal model and data analysis. Although we did not expect to find differences in responses of male versus female mice, we have examined the data by sex. Both age-matched male and female mice were used in experiments. Littermate CT mice were used in all experiments in which transgenic mice were used.
Animal study approval
All animal care and experimentation were conducted in France in compliance with the guidelines of the European Union (86/609/EEC) and the French Committee of Ethics (87/848) policies and with the specific approval from the local ministry-approved committee on ethics in animal experimentation (Ethics Committee UMS006 CEEA-122; project no. 21938-2019090417341270v5). Upon arrival in the animal facility, the animals were housed in accordance with European Directive 2010/63/EU. The mice were kept in cages at 20–24 °C with 45–65% humidity, in groups of three for females and two for males, and litters were housed in groups of five.
Timed pregnancies
W8 to W10 male and female mice were subjected to timed pregnancies and introduced to a breeding diet (SAFE A03; 69.2% cereals, 6% animal proteins, 20.2% plant proteins and 4.6% vitamins and minerals) at E0. Successful mating was judged by the presence of vaginal plugs, which defined E0.5 post-conception. Female weight was monitored all along the gestation period, and gestation was confirmed by a significant increase in body weight, where a weight gain of greater than 3.5 g from E0 to E13 was indicative of gestation. In addition to wild-type pairing, mating pairs were designed as follows: KitWsh/Wsh females with KitWsh/Wsh males, Kit+/Wsh females with Kit+/Wsh males and iDTAfl/fl females with Mcpt5–cre+ males.
Chronic PS model
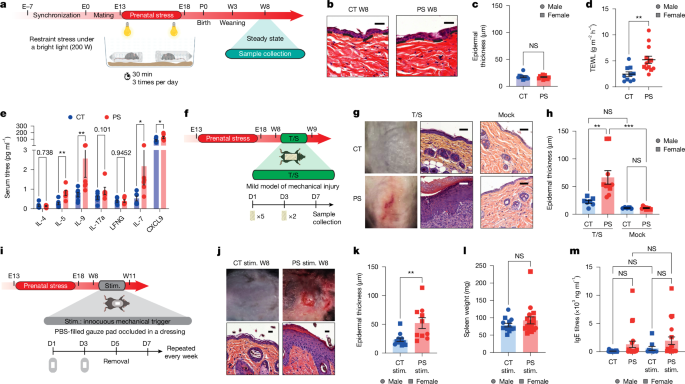
PS was induced, as previously described7. Briefly, pregnant dams were randomly attributed to CT and PS groups. The PS group was then subjected to chronic restraint stress in transparent drilled 50-ml Falcon Tubes (up to 6 mice per cage), put under a bright light (200 W), 3 times a day for 30 min each, from E13 until E18. The CT pregnant females were left undisturbed. Female dams were monitored every day during pregnancy and kept housed in groups of two or three animals until E18.5 after the last stress session when they were put in individual cages to prepare their nests. After birth, all pups were kept in the same cage with their biological mother. The pups were then weaned at the age of 21–28 days in distinct sex groups in new cages and introduced to a maintenance diet (SAFE A04; 84.1% cereals, 4% animal proteins, 8% plant proteins and 3.9% vitamins and minerals). Experiments were conducted at the desired age of offspring.
Metyrapone treatment
CT and PS dams were injected intraperitoneally with 50 µg g−1 of metyrapone (Bio-Techne), once on E11 and E12 and 30 min before each stress session from E13 to E18. The injection pattern and dose were determined to obtain an effective corticosterone synthesis inhibition while keeping the required levels for gestation. Serum for corticosterone dosage was collected immediately after the last session of stress on E18.5 and assessed for corticosterone concentrations.
In utero tamoxifen administration and fate mapping
For Cdh5–creERT2 fate mapping, ROSAmT/mG or ROSATdt/Tdt females were subjected to timed matings with Cdh5–creERT2+/− ROSAmT/mG or ROSATdt/Tdt, respectively. For induction of reporter recombination in the offspring, a single dose of 4OHT was delivered by intraperitoneal injections to pregnant females at E7.5 or E10.5. When the Cre is expressed in the Cdh5–creERT2;ROSATdt and Cdh5–creERT2;ROSAmT/mG mice, it excises the loxP-flanked stop cassette, allowing the expression of Tdt or the switching of the fluorescence from red (mT) to green (mG), respectively, permanently marking the Cre-expressing cells and their descendants. To counteract the adverse effects of 4OHT on pregnancies, 4OHT solutions were supplemented with progesterone. Females received a dose of 1.2-mg 4OHT and 0.6-mg progesterone. Pregnant dams were then subjected to PS, and offspring were euthanized at E18.5, W8 and W24. In cases when females could not give birth naturally, pups were delivered by C-section and cross fostered with lactating CD1 females.
Model of mild wet skin friction
Mouse back skin was shaved and treated with a PBS-filled gauze pad occluded in Tegaderm Transparent Dressing (3M Health Care) every 2 days. On day 5, the gauze pads were removed, and the mice were left without any treatment for 2 days. This cycle was repeated for extra 2 weeks. Two days after the last cycle of treatment, the mice were euthanized, and back skin specimens corresponding to the treated areas were obtained for analysis. Skin inflammation was graded by unbiased measurement of the epidermal thickness. Only skin lesions found in the patching area were taken into consideration.
Mild model of tape stripping and TEWL measurements
The back skins of W3 and W8–W12 mice were shaved using an electrical razor. To disrupt the skin barrier, tape strips (autoclave tapes) were pressed firmly against the shaved area and removed immediately after application. This procedure was repeated on day 1 (five tape strips for W8 and three tape strips for W3) and day 6 (two tape strips for W8 and three tape strips for W3). Tweezers were used to remove the tape in a fluent stroke. The tape strip application was always conducted by the same person to avoid any bias owing to strength and applied pressure differences. Barrier permeability was monitored using TEWL measurements (in g m−2 h−1) that were recorded in optimal room conditions (temperature 20 °C; 40–60% relative humidity). The Tewameter probe (wireless open chamber probe; Courage + Khazaka electronic) measures the density gradient of the water evaporation from the skin indirectly by two pairs of sensors (temperature and relative humidity) according to the diffusion law. The mice were euthanized on day 8, and CT versus tape-stripped skin were collected.
Multiplex proximity extension assay (Olink)
Skin sections were disrupted in RIPA Lysis Buffer (1:10; EMD Millipore Sigma-Aldrich) supplemented with protease inhibitor cocktail tablets (cOmplete Mini EDTA-free tablets; EASYpack; Roche) and homogenized using CKMix beads (Bertin) and Precellys 24 homogenizer. Skin sections were then processed with two cycles of 6,500 rpm for 20 s. The protein concentrations of clarified tissue were measured by Pierce BCA Protein Assay Kits (Thermo Fisher Scientific). Samples were subsequently normalized to a concentration of 0.5 mg ml−1 with the 1× RIPA Lysis Buffer.
The obtained skin lysates, amniotic fluid and serum samples were analysed using a proximity extension assay using the Olink Target 48 Mouse Cytokine panel in the presence of internal (incubation, extension and detection CTs) and external CTs. Data were analysed using a preprocessing normalization procedure. For each sample and data point, the corresponding quantification cycle (Cq) value for the extension CT was subtracted, thus normalizing for technical variation in one run. Normalization between runs was then performed for each assay by subtracting the corresponding ΔCq value for the median of the three calibrator replicates from the ΔCq values generated. The normalized protein expression unit generated was on the basis of a log2 scale. The protein concentration in standard concentration units (in pg ml−1) was obtained by fitting the normalized protein expression value to a standard curve. The results were reported in standard concentration units (in pg ml−1).
Mechanical alloknesis test
W8–W12 mice were shaved at the nape of the neck and acclimated for 30 min in the experimental room and then in Plexiglas boxes of the same size for 30 min, one day before the allokinesis test. The same habituation was carried out on the day of the test. Each mouse received five innocuous mechanical stimuli at the nape of the neck from two von Frey filaments (0.07 g and 0.4 g; Bioseb BIO-VF-M). The response of the mice after each stimulation was scored (0, no reaction; 1, head movement; 2, action towards the filament; 3, scratching), and the percentage of total response was plotted.
Sticky-tape assay
W8–W12 mice were acclimated for 30 min in the experimental room and then in Plexiglas boxes for 20 min. A white 12.7-mm circular adhesive tape was gently placed to the upper back of the mice between shoulders. The tape-directed responses of the mice were monitored as follows67: wet-dog shake, one bout; trying to reach tape with snout, one bout; burst of directed scratching with hind leg, one bout; neck grooming with front paw, one bout; grooming of head, lower back, paws, legs and ventral areas or tail, no bout. The recording was stopped when the mouse managed to get the tape off or after a trial time of 5 min. The following parameters were analysed: tape riddance (success/timeout), total number of bouts over the total trial time and bouts per minute, no response time (sum of time throughout which no bout occurred for 15 s or more) and time course of bouts represented as area under the curve of the time course plot (cumulative bouts versus time). Fisher’s exact test was used to test whether PS affects the rate of mice managing to get the tape off in time. Time courses of bouts were plotted per second and then averaged for each time point. The area under these averaged time course curves (baseline at 0 bouts) was calculated using the trapezoid rule for every Δx = 1 s using GraphPad Prism. The yielded averages ± s.e.m. and the averages of the total bouts and total bouts per minute were then statistically analysed by two-way ANOVA followed by Holm–Šidák multiple comparison tests.
von Frey filament test
W8–W12 were acclimated for 1 h in the testing arena. Increasing diameter filaments delivering increasing predetermined forces were applied perpendicularly to the hind paw surface of the mouse until they buckle. The 0.02-g von Frey filament was used as the starting filament. Subsequently, filaments of 0.04 g, 0.07 g, 0.16 g, 0.4 g, 0.6 g, 1.4 g and 2 g were used. Any brisk behaviour, including paw withdrawal, licking or shaking during the probing or immediately after the filament was removed, was considered as a positive response. For each mouse, the paw withdrawal threshold was determined as the filament force that induced a positive response on three of five trials.
Passive cutaneous anaphylaxis
The ears of W8 offspring were injected intradermally with 8-µg ciprofloxacine, an MrgprB2 agonist in 20-μl PBS. Ear swelling was measured every 10 min under light anaesthesia (2% isoflurane), starting just before injection (T0) and ending 2 h after injection (T120).
Peanut-induced anaphylaxis
Mice were sensitized with 1 mg of peanut extract (clinical-grade preparations used for skin testing; Stallergenes Laboratories) along with 10 μg of cholera toxin (Sigma-Aldrich) in 100-μl water (HCO3−) administered by means of oral gavage once a week for 4 weeks. One week after the last sensitization with peanut extract, the mice were challenged with the intraperitoneal injection of 1 mg of crude peanut extract (Greer Laboratories) in 100-μl PBS. Rectal temperature measurements were performed every 30 min, starting immediately before challenge (T0) and ending 2 h after challenge (T120).
Short model of asthma and lung dissociation
W8 offspring were sensitized by intranasal administration of lipopolysaccharide on day 0 (D0) and either PBS or 20 µg (30 µl) of the HDM strain Dermatophagoides pteronyssinus on D1 and D8, under general anaesthesia (not too long so that the mouse can breathe through its nose and inhale the liquid). On D11, the mice were euthanized, and lungs were collected and dissociated using mechanical and enzymatic dissociation. Briefly, lung lobes were collected in RPMI medium in Miltenyi C Tubes and then digested with 0.5 mg ml−1 of DNase I (Sigma-Aldrich) and 0.25 mg ml−1 of Liberase (Roche) using the Miltenyi gentleMACS (protocol: mouse lungs). Cell suspensions were then filtered, and red blood cells were lysed using ammonium chloride–potassium lysis buffer for 5 min. The pellet was recovered and stained for FACS analysis.
Resiniferatoxin treatment
To ablate TRPV1+ nociceptors, W4 CT or PS mice were subcutaneously injected with increasing RTX doses of 30, 70 and 100 μg kg−1 in 100-μl PBS for three consecutive days. Some mice were injected with similar volumes of dimethyl sulfoxide (DMSO) in 100-μl PBS. Four weeks later, denervation was assessed using the classical tail flick assay.
Chemical sympathectomy
6-OHDA (Sigma) was dissolved in vehicle solution (0.01% ascorbic acid) in saline solution. CT and PS mice received two intraperitoneal injections of 6-OHDA (150 mg kg−1) at 24-h interval. The mice were sacrificed 2 days after the first injection, and back skin was collected for imaging analysis.
Section preparation, histology, immunofluorescence and confocal microscopy
Mouse samples were either frozen in optimal cutting temperature compound (Tissue-Tek) or fixed in 10% formalin and embedded in paraffin. Frozen or paraffin-embedded mouse skin sections pretreated using a heat-induced epitope retrieval method (in 10 mM sodium citrate buffer (pH 6.0)) were permeabilized for 30 min in PBS 0.5% (w/v)% BSA and 0.3% Triton X-100 and incubated overnight at 4 °C with fluorophore-coupled antibodies or unconjugated antibodies: anti-Filaggrin (Ozyme; BLE905801; 1:50), anti-Loricrin (Ozyme; BLE905101; 1:50), anti-mouse Keratin 6A (Ozyme; BLE905701; 1:50), Anti-Claudin 1 antibody (Abcam; ab15098; 1:50), Alexa Fluor 647 anti-mouse CD45 (BioLegend; 103124; 1:400), anti-TH (Sigma; AB1542; 1:100), Alexa Fluor 647 anti-Tubulin β3 (BioLegend; 801210; 1:50), anti-human/mouse GFRα2 (R&D Systems; AF429; 1:200), anti-mouse NFH (Sigma; AB1989; 1:1,000), anti-GFP (Proteintech; pabg1; 1:200), anti-RFP (Takara; 632496; 1:200), Anti-Substance P (Merck Millipore; MAB356; 1:200) and anti-αSMA (Thermo Fisher Scientific; 14-9760-82; Clone 1A4; 1/100). The sections were then washed 3 times in PBS 0.5% (w/v)% BSA and 0.3% Triton X-100 and incubated, if needed, with the following secondary antibodies in PBS 0.5% (w/v)% BSA and 0.3% Triton X-100 for 2 h at room temperature in the dark: Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Invitrogen; A31571; 1:200), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (Invitrogen; A11012; 1:200), Donkey anti-Sheep IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (Invitrogen, A21099; 1:200), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Invitrogen; A11008; 1:200) and Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Thermo Fisher Scientific; A21247; 1:200). The slides were then washed, mounted in Mowiol medium and sealed with a coverslip. Z-stack images (512 × 512) were acquired using Leica STELLARIS, Leica SP8 and Zeiss LSM 710 Meta inverted confocal laser scanning microscopes and were processed using ImageJ (v.2.16.0) and Imaris (Bitplane; v.9.5.1) software. The MFIs of key epidermal proteins were analysed using the ‘measurement function’ of ImageJ software (v.2.16.0) on randomly chosen epidermal zones of identical areas on three non-overlapping sections (same total number of pixels). The cell bodies of neurons were counted on at least three images of three non-overlapping DRG sections (from lower thoracic to upper lumbar DRG) per mouse. Data were presented as percentage of Tdtomato+ neurons.
Haematoxylin and eosin
Mouse back skin sections were stained according to standard procedures at the histopathology department CREFRE U006. Slides were then scanned using a Pannoramic Digital Slide Scanner (3DHISTECH). For measurement of epidermal thickness, three randomly selected measurements of the distance between the stratum corneum and the basal layer of the epidermis were recorded using the slide viewing application CaseViewer 2.3.
Computational analysis of filament length of neurons in the skin
The 3D high-resolution images were taken using a Leica TCS SP8 MP Meta inverted confocal microscope, as described above. The images were then processed using the software Imaris (Bitplane; v.9.5.1). The filament tracer algorithm was applied in the Tubβ3, TH, IB4, NFH and Gfrα2 channels for precise tracing of the trajectories and shapes of Tubβ3+ (pan neuronal marker), TH+ (back skin, mainly c-LTMRs and can also stain sympathetic neurons), Tubβ3+ IB4+ neurons (back skin and non-peptidergic), NFH+ (glabrous skin and LTMRs) and Gfrα2+ (glabrous skin and non-peptidergic). Filament traces were then computed, and the sum of the filament lengths was calculated and normalized per square millimetre for all the samples.
Mouse tissue enzymatic digestion and gradient separation
Cell suspensions from embryonic and adult mouse tissues were obtained through mechanical dissociation and enzymatic digest. Briefly, adult back skin from W8 or W24 mice was collected in pre-digestion medium, finely minced and incubated at 37 °C on a rotating plate to remove epithelial cells and other impurities. The samples were then digested for 45 min on a rotating plate with 1.25 mg of Liberase (Sigma) and 2.5 mg of DNAse I (Sigma) to disaggregate the tissue. The samples were further dissociated with the Miltenyi gentleMACS Dissociator, and cells were then enriched with a Percoll gradient.
Fetal skin was collected, finely minced and digested in RPMI medium containing 0.1 mg ml−1 of DNAse I (Sigma), 400 U ml−1 of Collagenase I (Sigma), 0.1 mg ml−1 of Liberase DL (Roche) and 1 mg ml−1 of Dispase II (Roche) for 45 min on a rotating plate at 37 °C. The digested tissue samples were filtered.
Single-cell suspensions were then used for flow cytometry.
Flow cytometry
The cell suspensions were blocked with anti-mouse CD16/32 (S17011E; BioLegend) for 15 min at 4 °C. Surface staining was performed in FACS buffer for 1 h at 4 °C using the following antibodies: anti-mouse CD45 BV786 (BD Biosciences; 564225; 1:200), anti-mouse/human CD11b Brilliant Violet 605 (BioLegend; 101237; 1:200), anti-mouse CD117 SB436 (Thermo Fisher Scientific; 62-1171-82; 1:200), anti-mouse CD117 PE (BioLegend; 105807; 1:200), anti-mouse Ly6C Alexa Fluor 488 (BD Biosciences; 553104; 1:200), anti-mouse Ly6G APC Cy7 (BD Biosciences; 560600; 1:200), anti-mouse/human CD207 PE-Cy7 (BioLegend; 144209; 1:200), PerCP/Cyanine5.5 anti-mouse CD3 (BD Biosciences; 551163; 1:200), BV650 Anti-Mouse γδ T-Cell Receptor (BD Biosciences; 563993; 1:200), anti-mouse CD4 PE Cy7 (BD Biosciences; 552775; 1:200), Alexa Fluor 488 anti-mouse CD8 (BioLegend; 100726; 1:200), anti-mouse CD25 PE (BD Biosciences; 553075; 1:200), anti-mouse NK1.1 APC Cy7 (BD Biosciences; 560618; 1:200), anti-mouse Nkp46 BV605 (BioLegend; 137619; 1:200), anti-human/mouse KLRG1 BV421 (BioLegend; 138413; 1:200), anti-mouse CD11c PE CF594 (BD Biosciences; 562454; 1:200), anti-mouse MHCII AF700 (BioLegend; 107621; 1:200), anti-mouse CD24 PE Cy7 (BD Biosciences; 560536; 1:200), anti-mouse F4/80 BV711 (BioLegend; 123147; 1:200), anti-mouse CD170 (Siglec F) eFluor 660 (Thermo Fisher Scientific; 50-1702-82; 1:200), Alexa Fluor 647 anti-mouse CD64 (BD Biosciences; 558539; 1:200), anti-mouse ST2 APC (BioLegend; 146605; 1:200), anti-mouse FcERI PE (Miltenyi; 130-118-896; 1:200) and anti-mouse FcERI PE-Cy7 (BioLegend; 134317; 1:200). Data were acquired on a BD FACSymphony cytometer and were analysed using FlowJo (Tree Star; v.10.10.0) software. Cell sorting was performed on BD FACSAria.
Immune cell populations were gated as follows:
γδ T cells: CD45+CD11b−Ly6G−CD3+TCRγδ+
CD4+ T cells: CD45+CD11b−Ly6G−CD3+TCRγδ−CD4+CD8−
CD8+ T cells: CD45+CD11b−Ly6G−CD3+TCRγδ−CD4−CD8+
Regulatory T cells: CD45+CD11b−Ly6G−CD3+CD25+
Natural killer cells: CD45+CD11b−Ly6G−CD3−NK1.1+
Natural killer T cells: CD45+CD11b−Ly6G−CD3+NK1.1+
ILC2: CD45+CD11b−Ly6G−Nkp46−KLRG1+
ILC3: CD45+CD11b−Ly6G−Nkp46+KLRG1−
Dermal dendritic cell: CD45+CD11b+CD11c+MHC2+
Langerhans cell: CD45+CD11b+CD24+
Neutrophil: CD45+CD11b+Ly6C+Ly6G+
Monocyte: CD45+CD11b+Ly6C+Ly6G−
Macrophage: CD45+CD11b+F4/80+
Alveolar macrophage (adult lung): CD45+CD11c+Siglec-F+MertK+
Basophil: CD45+CD117−FceRI+
Eosinophil: CD45+CD11b+Siglec-F+
Mast cell (adult tissues): CD45+CD11b−CD117+FceRI+
Mast cell (fetal skin): CD45+F4/80−CD64−CD117+ST2+
All cells were pregated as viable (using SYTOX viability dye) singlets.
Tissue RNA extraction
Skin samples or DRG from adult mice were stored immediately in RNAlater then dissociated in TRIzol with the Miltenyi gentleMACS Dissociator. Tissue RNA was extracted using the RNeasy Qiagen Mini Kit according to the manufacturer’s guidelines. Fetal mast cells were sorted, as described above. RNA was then extracted using RNeasy Qiagen Micro Kit according to the manufacturer’s guidelines. RNA quality and titres were analysed using NanoDrop. RNA was either used for bulk RNA-seq or real-time quantitative polymerase chain reaction (qPCR).
Preparation of libraries for bulk RNA-seq
Sample quality CT and libraries were prepared at the GeT-Santé facility38,39. RNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The integrity of RNA was checked with a Fragment Analyzer (Agilent Technologies) using the RNA standard sensitivity kit. The 260/280 purity ratios were all greater than 1.8. Integrity indices revealed acceptable and homogeneous values for the RNA integrity number (8.2–8.6) and 28S/18S ratios (0.8–1.2). The RNA-seq paired-end libraries were prepared at the GeT-Santé facility according to Illumina’s protocol with some adjustments, using the TruSeq Stranded mRNA library prep Kit (Illumina). Briefly, messenger RNAs were first selected from either 3,000 ng (DRG sequencing) or 500 ng (sorted fetal mast cell sequencing) of total RNA using poly-T beads. RNAs were then fragmented for 2′ and retrotranscribed to generate double-stranded complementary DNA (cDNA). Compatible adaptors were ligated, allowing the barcoding of the samples with unique single indices. Eleven cycles of polymerase chain reaction (PCR) were applied to amplify libraries, and an extra double-sized purification step allowed to obtain 280–1,000 pb fragments. The quality of libraries was assessed using the HS NGS kit on the Fragment Analyzer (Agilent Technologies).
Bulk RNA-seq
The quantification and sequencing of libraries were performed at the GeT-PlaGe core facility (Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement). Libraries were quantified by means of qPCR using the KAPA Library Quantification Kit (Roche) to obtain an accurate quantification. Libraries were equimolarly pooled, and RNA-seq was then performed on one S prime lane of the Illumina NovaSeq 6000 instrument (Illumina) using the NovaSeq 6000 S prime v.1 Reagent Kit (300 cycles) and a paired-end 2 × 150-pb strategy.
FastQC was performed on sequence reads to evaluate the sequencing quality. Reads were then aligned on GRCm39 (mm10) as a reference genome using STAR (v.2.4.0)68 aligner and counted with HTseq (v.0.9.1)69, and a counting matrix was generated. Count matrix values were transformed using regularized log transformation implemented in DESeq2 (v.1.34.0)70. We performed data exploration using RStudio (v.2024.12.1).
Preparation of libraries for scRNA-seq
Approximately 16,000 cells per sample were encapsulated into droplets using Chromium Next GEM Single Cell 3′ Reagent Kits v.3.1 according to the manufacturer’s protocol (10x Genomics). Briefly, after generation of nanolitre-scale Gel bead-in-EMulsion (GEM) using Next GEM Chip G, GEMs were reverse transcribed in a C1000 Touch Thermal Cycler (Bio-Rad) programmed at 53 °C for 45 min and 85 °C for 5 min and held at 4 °C. After reverse transcription, single-cell droplets were broken, and cDNA was isolated and cleaned with Clean-Up Mix containing Dynabeads (Thermo Fisher Scientific). Then, cDNA was amplified with a C1000 Touch Thermal Cycler programmed at 98 °C for 3 min, 12 cycles of (98 °C for 15 s, 63 °C for 20 s and 72 °C for 1 min) and 72 °C for 1 min and held at 4 °C. Subsequently, the amplified cDNA was fragmented, end-repaired, A-tailed, index adaptor ligated and cleaned with Clean-Up Mix containing SPRIselect Reagent Kit (Beckman Coulter) in between steps. Post-ligation product was amplified and indexed with a C1000 Touch Thermal Cycler programmed at 98 °C for 45 s, 14 cycles of (98 °C for 20 s, 54 °C for 30 s and 72 °C for 20 s) and 72 °C for 1 min and held at 4 °C. The sequencing-ready libraries were cleaned up with SPRIselect beads.
Libraries prepared using 10x Genomics technology were pooled and charged with 1% PhiX on one S prime lane of the NovaSeq 6000 instrument (Illumina) using the NovaSeq 6000 S prime Reagent Kit v.1.5 (200 cycles) and the following sequencing parameters: 28 bp read 1, 8 bp index 1 (i7) and 150 bp read 2. The S prime lane generated a total of 434 × 106 reads.
Single-cell RNA sequencing analysis
Preprocessing
Transcript reads were demultiplexed, aligned to reference mouse genome (mm10 – GENCODE vM23/Ensembl 98) and quantified using the Cell Ranger v.7.0.1 (ref. 71). Quantified gene counts of the samples were preprocessed to remove ambient RNA decontamination using the decontX function from the celda package v.1.6.0 (ref. 72). The raw unfiltered matrix output of the Cell Ranger count of each sample was used as the respective background for the ambient RNA inference. Seurat package v.4.3.0.1 (ref. 73) was preferred as the main suite of analysis. Cells with more than 10% of their reads were assigned to mitochondrial genes, and cells with less than 100 detected genes were excluded from the dataset. After this quality control trimming, a total of 28,015 cells (7,211 cells from CT W8, 4,474 cells from PS W8, 6,812 cells from CT W24 and 9,004 cells from PS W24) were retained for further analysis.
Processing
We then ran the Seurat standard analysis pipeline: normalization, finding highly variable genes, scaling and PCA. Variance explainability of the principal component dimensions was observed, and the first 20 principal components were chosen for downstream analysis. The dataset consisted of two batches, which were integrated using the Harmony package v.1.0.3 (ref. 74), with the inclusion of the first 20 principal component dimensions. The k-nearest neighbour graph, UMAP and t-distributed stochastic neighbour embedding projections were computed on the basis of the first 20 corrected principal components. To scrutinize the distinct communities within the dataset, unsupervised clustering was performed using the Louvain algorithm with a resolution of 0.3.
Cell identification
FindAllMarkers() function was used to determine the marker genes of the computed unsupervised clusters of the cells. By examining the expression of a set of canonical markers within our clusters, we successfully identified a total of 13 distinct cell types: Cd3e, Itgae and Trdc identifying γδ T cells; Cd207 and Ly75 identifying Langerhans cells; Xcr1 and Flt3 identifying dermal dendritic cells; Lyz2 and Mrc1 identifying conventional macrophages; Gzmb and Ncr1 identifying natural killer cells; Ms4a1 and Cd79b identifying B cells; S100a9 and S100a8 identifying neutrophils; Il17a and Il23r identifying ILC3s; Ccr6 and Gata3 identifying ILC2s; Cpa3 and Tpsb2 identifying mast cells; and Krt14 identifying keratinocytes.
Differential gene expression analysis
For the annotated cell types, the Poisson test within Seurat’s FindMarkers() function was run to compare one group to another. Genes with Padj < 0.05 and an absolute average log2(fold change) greater than 0.58 were determined to be significant in the given DEG test.
Preparation of libraries for single-nucleus Multiome ATAC and Gene Expression
The back skin of W8 and W24, CT and PS offspring (n = 4) was processed, and single-cell suspensions were enriched in CD45+ immune cells.
Nuclei isolation
Single nuclei were isolated following the 10x Genomics Nuclei Isolation for Single Nuclei Multiome ATAC + Gene Expression Sequencing protocol (CG000365-Rev C). Briefly, fresh cells were counted using a Countess 3 Automated Cell Counter (Thermo Fisher Scientific), and 150,000 cells were used. The cell pellet was homogenized in 100 µl of chilled 0.025% IGEPAL lysis buffer (10x Genomics). Lysate was incubated on ice for 2 min. At the lysis end, 1 ml of chilled wash buffer (10x Genomics) was added. After centrifugation (500 relative centrifugal force; 5 min; 4 °C), the supernatant was carefully removed, and the nuclei were resuspended in 1 ml of wash buffer. The nuclei suspension was then filtered through a 40-μm strainer and centrifuged (500 relative centrifugal force; 5 min; 4 °C) for a total of three washes to remove debris. Depending on nuclei number, the pellet was resuspended in between 3.4 and 15.5 µl of diluted nuclei buffer (10x Genomics) to have 8,060 nuclei per microlitre.
Single-nucleus partitioning and library preparation
Nuclei were immediately treated using the 10x Genomics Chromium Next GEM Single Nuclei Multiome ATAC + Gene Expression protocol (CG000338-Rev F). After transposition, the nuclei were diluted to optimal concentration and loaded into the Chromium Next GEM Chip J, followed by partitioning into GEMs using the Chromium X (10x Genomics). After GEM rupture, cDNA and transposed DNA fragments were purified using Dynabeads MyOne Silane (Thermo Fisher Scientific). Transposed DNA fragments underwent seven PCR cycle amplification to generate chromatin accessibility libraries, whereas cDNA fragments were further processed through end-repair, adaptor ligation and ten indexing PCR cycles to generate enriched gene expression libraries.
Sequencing
The resulting gene expression and ATAC libraries were quantified using a Qubit Fluorometer (Thermo Fisher Scientific) and analysed for fragment size distribution using an Agilent TapeStation before sequencing. The libraries were pooled at the recommended ratio and sequenced on an Illumina NovaSeq 6000 platform at the Technology Cluster of the Toulouse Cancer Research Centre (CRCT) using paired-end reads (read 1, 50 bp; read 2, 49 bp; index 1, 8 bp; index 2, 24 bp) and using paired-end reads (read 1, 28 bp; read 2, 90 bp; index 1, 10 bp; index 2, 10 bp), targeting a sequencing depth of approximately 50,000 read pairs per nucleus for gene expression to achieve a sequencing depth of 3,350 median fragments per cell for 19,946 cells post-aggregation.
Multiome data processing and integration
Preprocessing and quality control
Single-nucleus multiome sequencing data, including ATAC-seq and single-nucleus RNA-seq reads, were processed using Cell Ranger ARC (v.2.0.2). Reads were demultiplexed, aligned to the reference mouse genome (mm10 – GENCODE vM23/Ensembl 98) and quantified to generate filtered feature matrices and ATAC fragment files. The resulting datasets were loaded into Seurat (v.5.1.0) and Signac (v.1.14.0) for downstream analysis. Raw read files from all samples were merged into a single Seurat object, followed by quality control filtering to remove low-quality or multiple nuclei. Nuclei were retained if they met the following criteria: (single-nucleus RNA sequencing) 300 or more RNA features, total RNA counts of less than 5,000 and less than 10% mitochondrial RNA content; (snATAC-seq) ATAC peak counts between 100 and 20,000, nucleosome signal less than 0.75 and transcription start site enrichment score above 2. After filtering, a total of 12,499 high-quality nuclei were retained for downstream analysis: 2,130 nuclei from CT W8, 4,094 nuclei from PS W8, 3,767 nuclei from CT W24 and 2,508 nuclei from PS W24.
Single-nucleus RNA sequencing data processing
Gene expression values were log-normalized and scaled. Highly variable genes were identified; PCA was performed, retaining 50 principal components. Batch correction was applied using Harmony, and the corrected embeddings were stored for downstream analysis.
snATAC-seq data processing
For chromatin accessibility data, peaks with a minimum of five reads were retained. Term frequency–inverse document frequency normalization was applied, followed by singular value decomposition, keeping 50 latent semantic indexing components. Batch correction was applied using Harmony, and corrected embeddings were stored.
Multiomic integration and clustering
A weighted nearest neighbour graph was constructed using Harmony-corrected embeddings from RNA and ATAC data. The first 50 dimensions from RNA and 2–50 dimensions from ATAC were used. A joint UMAP was generated, and clustering was performed using the Louvain algorithm with a resolution of 0.1. For cell identification, similar markers to those listed above were used.
Differential analysis of accessible regions
To identify DARs of chromatin, pairwise comparisons were performed across experimental conditions. The analysis was conducted using the FindMarkers() function in Seurat with the model-based analysis of single-cell transcriptomics test, considering the chromatin accessibility data (ATAC assay). Comparisons were investigated for PS W8 versus CT W8, PS W24 versus CT W24 and PS W24 versus PS W8 groups. Only regions with a minimum detection rate of 5% across cells were considered. Genes associated with DARs were annotated using the ClosestFeature() function of Signac package (v.1.14.0). Significant DAR-associated genes were defined on the basis of a false discovery rate threshold of 0.05 and a log2(fold-change) cutoff of 1.5. Summary statistics were recorded for each comparison, and the results were visualized using dot plots displaying the number of genes associated with DARs.
Real-time qPCR analysis
RNA (1 μg) preparations from mouse skin were used for total RNA reverse transcription with SuperScript III First-Strand Synthesis System kit (Invitrogen) using random hexamers (Thermo Fisher Scientific) for priming. Transcripts encoding chemokine (C–C motif) ligand 20, thymic stromal lymphopoietin, interferon-γ, ILs (IL-10, IL-12p35, IL-12p40, IL-13, IL-17A, IL-17F, IL-22, IL-4, IL-5 and IL-6), oncostatin M, retinoic acid receptor-related orphan nuclear receptor-γt, signal transducer and activator of transcription (Stat; Stat3, Stat4 and Stat6) and transforming growth factor-β1 were quantified using real-time PCR using specific forward and reverse primers, the Takyon SYBR 2X MasterMix blue dTTP kit (Eurogentec) and quantified using the LightCycler 480 II (Roche Diagnostics).
Total IgE and corticosterone enzyme-linked immunosorbent assay
Blood samples were collected and centrifuged at 15,000g for 5 min. Sera were then collected and stored at −20 °C until use. Total IgE titres and corticosterone concentrations were quantified using an enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen and Abcam, respectively) following the manufacturer’s recommendations.
Single-cell analysis of mast cell degranulation dynamics
Mast cells from E18.5 fetal skin (n = 24) were sorted and plated into poly-d-lysine-coated (5 µg ml−1; Sigma-Aldrich) six-well glass bottom plates with coverglass (IBL Baustoff + Labor) in Tyrode’s buffer at 37 °C for 30 min, supplemented with 5 µg ml−1 avidin–sulforhodamine 101 (avidin SRho; Sigma-Aldrich). Cells were then stimulated with vehicle (Tyrode ± chloroform (negative CT)), 100 µM substance P (positive CT), 100 ng ml−1 of corticosterone and amniotic fluid (from CT, PS, CT-Met and PS-Met yolk sacs). Fluorescence was recorded after 30 min in a controlled atmosphere (using a ZEISS stage incubation system with objective heater; 37 °C) using Leica Sp8 confocal laser scanning microscope. MFI was quantified using the measurement function of ImageJ software for each defined single cell.
DRG dissociation, culture and calcium imaging
DRG from all spinal levels were collected in Hanks’ balanced salt solution without Ca and Mg and were digested with a mixture of 5 mg ml−1 of dispase and 1 mg ml−1 of collagenase type I at 37 °C for 45 min. DRG were then washed and further triturated with increasing gauge syringes in DMEM supplemented with 0.15 mg ml−1 of DNAse I. After dissociation, cells were spun at 300g and resuspended in medium before being plated in Nunc Lab-Tek II CC2 Chamber Slides with cover (Thermo Fisher Scientific). DRG were cultured for 24 h in DMEM supplemented with 50 ng ml−1 of nerve growth factor and 1:50 NeuroCult (STEMCELL) at 37 °C. The cells were loaded with Fluo-4 for 30 min in the dark at 37 °C in HEPES/Hanks’ balanced salt solution immediately before imaging. The cells were imaged for 20 s to establish a baseline. DRG were then stimulated with 100 µM β-alanine, 10 nM capsaicin, 2 nM MRS2365 or 400 ng ml−1 of corticosterone. At the end of every imaging trial, 50 mM KCl was added as a positive CT. Damaged, detached, high-baseline and motion-activated cells were excluded from analysis.
Pregnant women cohort and cortisol ELISA
This study was registered with the identifier NCT05207059 at ClinicalTrials.gov37. All subjects were recruited after obtaining necessary ethical approvals from the SingHealth Centralised Institutional Review Board in Singapore, and informed consent was obtained for every participant. Eligible participants were women aged 21–40 years with a body mass index of 25–40 kg m−2; intending to reside in Singapore for the next 4 years; identifying as Chinese, Malay, Indian or a combination of these ethnic groups; and planning to conceive within a year. Exclusion criteria included current pregnancy; known type 1 or type 2 diabetes; and recent use (past month) of anticonvulsants, oral steroids, certain contraceptives, fertility treatments or medications for human immunodeficiency virus or hepatitis B/C. Maternal blood was collected from pregnant women at two distinct time points: 6–10 gestational weeks and during delivery. The freshly processed blood was used to evaluate basophil reactivity using the BAT. BAT was performed, as described previously75, using HDM allergen, and responders were defined as those exhibiting greater than 38% degranulation upon HDM stimulation. Cortisol measurements in plasma were performed using ELISA kit (Abcam) following the manufacturer’s recommendations.
Statistics
Statistical tests were performed with the software Prism 8 (GraphPad software v.10.4.2). Kruskal–Wallis test for multiple comparisons using Dunn’s correction, two-way ANOVA for multiple comparisons using Šidák corrections, Fisher’s exact test and Mann–Whitney U-test were performed on samples, as noted in the respective figure legends. All statistical tests were two-sided. A P value of less than 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.