Syntheses of EVT0185 and its mono-CoA derivative
EVT0185 and EVT0185-CoA were synthesized at Symeres (www.symeres.com); experimental methods and compound characterization are presented in the Supplementary Information.
Mouse models
Animal experiments were carried out using the guidelines approved by the Animal Research Ethics Board at McMaster University (Steinberg Laboratory Animal Utilization Protocol #16-12-42, 21-01-04) or the Institutional Animal Care and Use Committee (IACUC) at Icahn School of Medicine at Mount Sinai (IACUC approval# PROTO202100080). The tumour development was monitored based on the established progression timelines for mouse HCC models. The welfare of the animals was evaluated through observation of visible masses, impaired mobility and marked weight loss. However, no specific tumour volume end points were defined for intrahepatic tumours. All experimental procedures did not exceed the limits set by the Animal Utilization Protocol or IACUC.
WD-DEN mouse model
C57BL/6J male mice were housed within the McMaster University Central Animal Facility. At 2 weeks of age, mice were injected with DEN at 25 mg kg−1 body weight. After weaning, mice were housed with littermates and fed a normal chow diet (8640 22/5, Teklad). At 10 weeks of age, mice were maintained on chow diet (control) or switched to a high-fat and high-fructose diet (WD), which consisted of 40% kcal fat (mostly palm oil), 20% kcal fructose and 0.02% wt cholesterol (D19101102, Research Diets) and housed at thermoneutral conditions (26–29 oC)26. Throughout the experiments, mice were housed in ventilated cage racks with ad libitum access to food and water. An automatic timing device was used to maintain an alternating 12-h cycle of light and dark. After 29 weeks, mice were euthanized, and blood and tissues were collected.
-
(1)
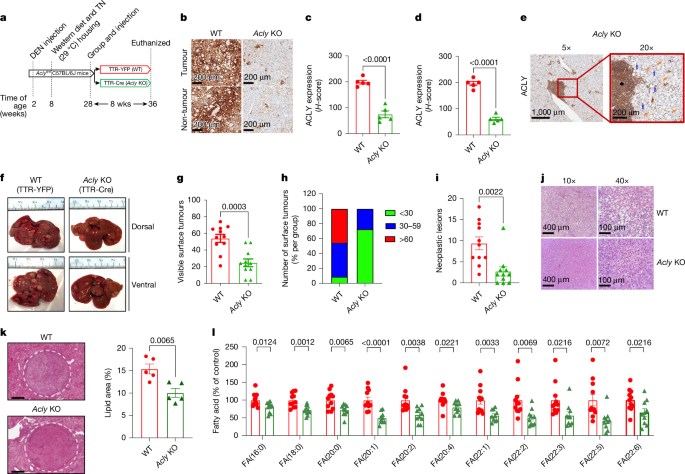
For the WD-DEN Acly-KO mouse model (Figs. 1 and 5), at 2 weeks of age, C57BL-6/Aclyfl/fl mice (obtained from JAX) were injected with DEN at 25 mg kg−1 body weight. After weaning, mice were housed with littermates and fed a normal chow diet (8640 22/5, Teklad). At 10 weeks of age, mice were switched to a high-fat and high-fructose diet (WD): 40% kcal fat (mostly palm oil), 20% kcal fructose and 0.02% cholesterol (D19101102, Research Diets) and housed at thermoneutral conditions. When the mice reached 7.5 months, they were bled from the tail vein, and plasma AFP levels were determined. Animals were grouped based on their AFP levels, so there were no differences between groups, and were then injected with AAV intravenously (AAV8-YFP or AAV8-TTR-Cre) as previously described26. After 4 (early timepoint) or 8 (late timepoint) weeks following AAV injection, mice were anaesthetized using ketamine–xylazine, and terminal blood was collected by cardiac puncture. For the B cell depletion experiments, AFP levels were assessed as described above after 7.5 months, and mice were randomized before being injected with AAV intravenously (AAV8-YFP or AAV8-TTR-Cre) and intraperitoneally injected with 250 µg of isotype control (400566, BioLegend) or anti-CD20 antibody (152104, BioLegend) with tissues collected 8 weeks later.
-
(2)
For the WD-DEN EVT0185 and bempedoic acid study (Fig. 3), male mice housed within the McMaster University Central Animal Facility were used for these experiments. C57BL/6J mice were injected with DEN at 2 weeks of age and followed the above-mentioned protocol. Animals were grouped based on their AFP levels and orally gavaged with a single daily dose of vehicle (1.5% CMC and 0.2% Tween-20), EVT0185-30 or 100 mg kg−1 or bempedoic acid-100 mg kg−1. After 4 weeks of treatment, mice were anaesthetized using ketamine–xylazine, and terminal blood and tissues were collected.
WD-CCl4 HCC model
Male C57BL/6J mice obtained from Jackson Laboratory were used for these experiments. Mice at 6–8 weeks of age were fed ad libitum with WD (Envigo; TD.120528, Teklad Custom Research Diet) and maintained on sugar water (fructose (23.1 g l−1) + glucose (18.9 g l−1)) for the duration of the study. Animals received weekly intraperitoneal injections of CCl4 (0.2 μl, 100% CCl4 per gram of body weight) throughout the study as described below.
The prevention study was conducted after 12 weeks of CCl4 injections + WD at McMaster University with mice orally gavaged with vehicle or EVT0185 (100 mg kg−1) for 18 weeks.
The combination treatment study was conducted after 19 weeks of CCl4 injection + WD at Mont Sinai with mice orally gavaged with EVT0185 (50 or 100 mg kg−1), sorafenib p-tosylate (sorafenib; at 15 mg kg−1; S-8502, LC Laboratories), lenvatinib mesylate (lenvatinib; 7 mg kg−1; 29832, LC Laboratories) or sorafenib + EVT0185 (100 mg kg−1) or lenvatinib + EVT0185 (100 mg kg−1).
The combination study with immunotherapy was conducted after 24 weeks of CCl4 injections + WD at McMaster University with mice being injected with InVivoPlus anti-mouse PDL1 (B7-81; BP0101), and InVivoPlus anti-mouse VEGFR2 (DC101; BP0060) alone or in combination with EVT0185 (100 mg kg−1). Isotype controls received InVivoPlus rat IgG1 isotype control, horseradish peroxidase (HRP; BP0088) and InVivoPlus rat IgG2b isotype control, anti-keyhole limpet haemocyanin (LTF-2; BP0090) as a single intraperitoneal injection containing 200 µg of each antibody in a total volume of 100 µl. Antibodies were prepared in InVivoPure pH 7.0 Dilution Buffer (IP0070). After 6 weeks of treatment, animals were anaesthetized using ketamine–xylazine, and terminal blood was collected by cardiac puncture.
WD-HCC mouse model
Male C57BL/6J mice obtained from Jackson Laboratory were used for these experiments. Mice at 6 weeks of age were fed ad libitum with WD, which consisted of 40% kcal fat (mostly palm oil), 20% kcal fructose and 0.02% wt cholesterol (D19101102, Research Diets). Throughout the experiments, mice were housed at thermoneutral conditions (26–29 °C). An automatic timing device was used to maintain an alternating 12-h cycle of light and dark. After 18 months, AFP-positive mice were divided into two groups and received a single daily dose via gavage of either vehicle (1.5% CMC and 0.2% Tween-20) or EVT0185 (100 mg kg−1). After 4 weeks of treatment, mice were anaesthetized using ketamine–xylazine, and terminal blood and tissues were collected. Blood samples were centrifuged at 10,000 rpm for 10 min at 4 °C and serum was collected and stored at −80 °C freezer. The liver was weighed, a photo was taken and the number of visible lesions on the surface of the liver was counted and recorded. A portion of tumour-free liver (approximately 200 mg, caudate lobe) and tumours were removed, flash-frozen in liquid nitrogen and stored for molecular analysis. The remainder of the liver was placed in 10% formalin, fixed for 48 h and stored in 70% ethanol until it was embedded in paraffin, sectioned and mounted on slides for histological analysis.
Orthotopic liver cancer model
1 day before surgery, male NOD-Rag1nullIL2rgnull (NRG) received intraperitoneal injections of 100 mg kg−1 cyclophosphamide and subcutaneous injections of 5 mg kg−1 carprofen. One hour before surgery, Hep3B-Luc cells were resuspended on ice in a 1:1 dilution of phenol red-free Matrigel (356237, Corning) and cold PBS (10010023, Gibco), aliquoted in 50 µl volumes at a cell density of 2 × 107 cells per millilitre. During surgery, mice were anaesthetized with 2% isoflurane (1001936040, Baxter), hair removed with 3-in-1 hair removal lotion (061700222611, Nair) and placed in a supine position on a heating pad, with nose fitted in an anaesthesia nose cone. The abdomen was disinfected and an approximately 2-cm horizontal incision was made below the left coastal margin from the midline of the abdomen. Separating the skin from the peritoneum, a second and smaller horizontal incision was made across the peritoneum to expose the liver. Next, the left lobe of the liver was withdrawn, stabilized and tumour cells were injected into the liver parenchyma while applying pressure using a cotton tipped applicator (4305, Dynarex) to enable haemostasis. Following injection, the liver was returned into the peritoneal cavity and the peritoneum was closed with multiple single interrupted sutures using 4-0 Vicryl sutures (J743D, Ethicon). The skin was closed using 9-mm wound clips (RS-9262, Roboz). Finally, mice received post-surgical subcutaneous injections of 5 mg kg−1 carprofen and were placed in cages on a heating pad containing diet recovery gel (72-06-5022, Clear H2O) and monitored until they were ambulatory. Mice were monitored for recovery for 7 days after surgery. Wound clips were removed on day 6, and mice were imaged for tumour progression on day 7 and weekly thereafter. Induction of in vivo short hairpin RNA (shRNA) expression was achieved via supplementing drinking water with 2 mg ml−1 doxycycline (Dox; D9891, Sigma-Aldrich) and 5% sucrose starting on day 7 post-surgery and every 2–3 days thereafter. In parallel, control mice were fed with drinking water supplemented with 5% sucrose without Dox. Tumour progression was followed until the end point, which is defined as the loss of more than 20% of the body weight. Mice were culled collectively when the first mouse in the experimental cohort (Hep3B-shACLY-Luc) reached the end point. Liver and tumour tissues were weighed and collected for downstream analysis. Tumour progression was monitored via bioluminescence imaging using the IVIS Spectrum In Vivo Imaging System (124262, Perkin Elmer). Bioluminescence was achieved through intraperitoneal injection of d-luciferin dissolved in saline (S8776, Sigma-Aldrich) at a concentration of 0.15 mg g−1 of body weight. IVIS imaging was performed on the auto-exposure setting at 10-min post-injection.
DNL assays
Compound screening
Primary mouse hepatocytes were isolated from C57BL/6J mice as previously described55 and seeded in white opaque 96-well plates, and the next day were serum starved for 2 h followed by the treatment with EVT compounds (0, 0.1, 0.3, 0.5, 1, 3, 10, 30, 60 and 100 µM) in the presence of 14C-acetate (1 µCi ml−1) in a concentration-dependent manner. After 4 h of treatment, plates were washed two times with 1× PBS and 100 µl microscint fluid (Microscint O, part #601361) was added to each well. Plates were wrapped with aluminium foil and were shaken at 250 rpm for 2 h. After 2 h of shaking, 14C incorporation into the lipid fraction was determined by liquid scintillation counting using a TopCount NXT Microplate Scintillation and Luminescence Counter (Perkin Elmer).
To compare the effect of bempedoic acid and EVT0185 in WT and Acly-KO mice, Aclyfl/fl mice were injected with hepatocyte-targeted AAV8-TTR expressing either YFP (WT) or Cre recombinase (Acly KO) via the tail vein to induce Acly genetic deletion as previously described26. After 2 weeks, hepatocytes from each mouse were collected and de novo lipogenesis (DNL) was assessed as previously described26. In brief, the cells were resuspended in complete William’s Media E and plated in six-well plates and allowed to adhere for 4 h. Cells were then washed with warm PBS and switched to fresh fetal bovine serum (FBS)-free (serum-free) William’s Media E for 2 h. Following a 2-h serum starvation, cells were washed in warm PBS, then treated with serum-free media supplemented with 1 µCi ml−1 [14C]-lactate (NEC599050UC, Perkin-Elmer) or 0.5 µCi ml−1 [14C]-acetate (NEC553050UC, Perkin-Elmer) for 18 h in the presence of 1 µM of EVT0185 or bempedoic acid both of which were dissolved in DMSO. After incubation, cells were washed three times with ice-cold PBS, scraped with 1 M KOH/EtOH, and incubated for 2 h at 70 °C with gentle agitation. After cooling, a 1:2 mixture of H2O:n-hexane was added to each sample, vortexed and centrifuged for 5 min (1,500 rpm). The top phase was transferred to a scintillation vial combined with Ultima Gold (NC0169557, Revvity) for counting of the sterol fraction. 2 N HCl and petroleum ether were added to the remaining bottom phase, which was vortexed and centrifuged for 5 min (1,500 rpm) at room temperature. The top phase was transferred to a scintillation vial for counting of the fatty acid fraction using a TopCount NXT Microplate Scintillation and Luminescence Counter (Perkin Elmer).
In vivo assessment of liver DNL
Male C57BL/6J mice (8 weeks of age) were obtained from Jackson Laboratory and were fed a normal chow diet upon arrival. At approximately 10 weeks of age, mice were given a diet containing high-fat and high-fructose (rodent diet with 40% kcal fat (mostly palm oil), 20% kcal fructose and 0.02% cholesterol (D19101102, Research Diets) and housed at thermoneutral conditions (26–29 °C). After 7–8 months, the mice were divided into four groups and received a single daily dose via gavage of either vehicle (1.5% CMC and 0.2% Tween-20) or EVT0185 at a dose of 10, 30 or 60 mg kg−1 for 7 days. On the morning of day 8, animals received a final dose, and after 1 h, 14C glucose (PerkinElmer) was administered at a concentration of 12 μCi per mouse in a volume of 0.1 ml in 0.9% saline (intraperitoneal). One hour after 14C glucose was given, animals were anaesthetized by intraperitoneal injection of ketamine–xylazine (150 mg and 12.4 mg kg−1, respectively). Blood was drawn through cardiac puncture; the liver was removed, and a sample from the left lobe was frozen in liquid nitrogen. Liver tissue was chipped on dry ice and the weight of the chip was weight recorded (30–50 mg of tissue). Liver tissue was homogenized in 1 ml of 2:1 chloroform:methanol using a bead homogenizer at 5,000 rpm for 2 × 12 s. Samples were incubated with gentle shaking at 4 °C for 2 h, vortexed for 2 × 12 s, and then centrifuged at 7,000 rpm for 10 min at 4 °C. The supernatant was transferred to a 1.5-ml Eppendorf tube and 200 μl of 0.9% saline was added. Samples were vortexed for 2 × 12 s and centrifuged at 3,000 rpm for 10 min at 4 °C. Next, 200 μl of the lower organic phase was removed and added to 5 ml of scintillation fluid. The amount of radioactivity in the sample was measured by scintillation counting. The number of disintegrations per minute was determined over 5 min and normalized to the amount of liver tissue. Of plasma obtained at termination, 5–10 µl was also counted and lipid per gram of tissue counts were normalized to plasma counts.
Measurement of respiratory exchange ratio by indirect calorimetry
Male C57BL/6J mice were fasted overnight (approximately 12 h) until food and 30% fructose water were again made accessible the next day at 7:30. Animals were allowed to feed ad libitum for approximately 2 h. The respiratory exchange ratio (RER) was monitored in metabolic cages using Oxymax/CLAMS (Comprehensive Laboratory Animal Monitoring System) equipment and software (Columbus Instruments). The calorimetry system consists of metabolic cages each equipped with water bottles and food hoppers connected to monitor food intake. All animals had ad libitum access to standard rodent chow and water throughout the study. Those animals with RER > 1 were administered with or without EVT0185 at a dose of 100 mg kg−1 via gavage. Animals remained in the metabolic cages and RER was monitored for 24 h.
Analytical methods
AFP ELISA
Plasma AFP levels were determined using a mouse AFP Duoset ELISA kit (DY5369-05, R&D Systems) following the manufacturer’s protocol. In brief, 100 µl sample (1:1,000 in reagent diluent) was added to each well of a 96-well plate coated with mouse AFP capture antibody. Next, the plates were washed with wash buffer and 100 µl detection antibody was added to each well, followed by the Streptavidin HRP and substrate solution. The reaction was stopped by stop solution and the optical density of each well was measured in a microplate reader set to 450 nm.
Histological analysis
Following fixation with 10% neutral-buffered formalin for 48 h, the cassettes with a medial lobe of the liver were switched to 70% ethanol. Livers were then processed, paraffin-embedded, serially sectioned and stained with haematoxylin and eosin (H&E), antibodies targeted to CD3+ and CD19+ cells by the McMaster Immunology Research Centre Histology Core Facility. Images were taken using a Nikon 90i Eclipse upright microscope at indicated magnifications.
Liver histology scores were assigned to liver sections by a pathologist who was blinded to the treatment conditions. Steatosis, inflammation and hepatocellular ballooning scores were assigned from H&E-stained liver sections as described by Kleiner et al.56. NAFLD activity scores (NAS) were obtained from the sum of these three scores. The surface area of tumour in the liver was quantified using qupath software from H&E-stained slides. The quantification of tumour CD3 and CD19-positive staining was completed from the same histological sections using HALO software by an analyst who was blinded to the treatment conditions.
The H-score system was used for ACLY immunohistochemistry evaluation on tumoural and non-tumoural hepatic tissues. This was determined by the multiplication of the percentage of cells with cytoplasmic staining intensity ordinal value (0 for no, 1 for weak, 2 for medium and 3 for strong staining), which ranges from 0 to 300 possible values. The percentage of each staining intensity was taken into account by the eye and multiplied by the staining intensity. The whole tumoural areas of each liver were evaluated in ×10 magnification as well as non-tumoural areas, and H-scores were calculated separately.
$$H-{\rm{score}}=( \% \,{\rm{weak}}\;{\rm{staining}})(1)+( \% \,{\rm{medium}}\;{\rm{staining}})(2)+( \% \,{\rm{heavy}}\;{\rm{staining}})(3)$$
Hepatic lesions were characterized into neoplastic and non-neoplastic proliferative lesions by a histopathologist according to the International Harmonization of Nomenclature and Diagnostic Criteria for Lesions in Rats and Mice (INHAND) guidelines57. All lesions had a hepatocellular phenotype. Non-neoplastic proliferative lesions, including foci of altered hepatocytes (FAHs), are referred to as premalignant lesions because they are composed of phenotypically altered cells demonstrating a higher risk of progression to malignancy than normal cells58,59. FAHs were defined as sharply demarcated circular, oval or irregularly shaped foci with preserved hepatic architecture and without compression of surrounding liver parenchyma. The basophilic-type FAH was the dominant type observed in this study, composed of slightly smaller hepatocytes with basophilic cytoplasm and slightly enlarged pleomorphic nuclei with prominent nucleoli, showing increased mitotic activity compared with background hepatic parenchyma. Scattered eosinophilic FAHs were also identified, composed of enlarged, polygonal hepatocytes with distinctly granular and pale pink, intensely eosinophilic or ground-glass appearance, as well as few clear cells and mixed cell types of FAHs. Neoplastic lesions as HCCs were characterized by loss of normal architecture (thickened hepatocytes plates), revealing mostly solid patterns as well as focal pseudoglandular and trabecular (micro or macro) patterns, including unpaired arteries or arterioles, consist of polygonal cells with more nuclear atypia (irregular nuclear membrane, pleomorphism, increased nuclear-to-cytoplasmic ratio, multinucleation and prominent nuclei), higher mitotic rate and cytoplasmic alterations include Mallory–Denk bodies and hyaline bodies57.
Transcriptome analysis
Bulk RNA-seq analysis
RNA isolation
Frozen tumour tissue (approximately 30 mg) was homogenized with Buffer RLT (RNeasy Mini Kit, 74106, Qiagen), and RNA isolation was performed using the RNeasy kit (74106, Qiagen) following the manufacturer’s instructions. Sample quality was assessed using the Agilent 2100 Bioanalyzer G2938C with the Aligent RNA 6000 Nano Kit (Agilent).
RNA-seq differential gene expression analysis
Only samples passing the RNA integrity number threshold of 7.0 were used for sequencing. mRNA was enriched using the NEBNext Poly(A) mRNA Isolation Module (NEB) followed by library preparation using the NEBNext Ultra II Directional RNA library kit (NEB). Next-generation sequencing was conducted at the McMaster Genomics Facility, Farncombe Institute, McMaster University, using Illumina HiSeq 1500 (Illumina). Samples were randomly distributed across lanes of a HiSeq Rapid v2 flow cell to eliminate lane-specific effects, and single-end 50-bp reads were generated at 12.5 million clusters per sample. Sequence quality was assessed using FastQC followed by the removal of low-quality reads and adapter sequences using Cutadapt. Genome alignment was performed using HISAT2 and the Mus musculus mm10 reference genome. Quantification of reads was performed using feature Counts.
Comparison of mouse models
We obtained raw RNA-seq files for tumour samples derived from control-DEN and WD-CCl4 models from PRJNA488497 (ref. 28) and PRJNA386995 (ref. 29), respectively. Sequencing files were processed using the same method as described for in-house samples. Count-level expression data were adjusted for surrogate batch effect across all samples using ComBat-Seq as implemented in the sva package in R. Subsequently, adjusted expression data were normalized to transcript per million. Molecular classification of Hoshida subtypes for human sample and mouse models were performed using the nearest template prediction module as implemented in the Gene Pattern platform online. Pairwise correlation between each mouse sample and each human sample were averaged on a per-model basis for mouse models, and on a per-subtype basis for human samples. Consistent with a previous report15, we validated the unique transcriptomic enrichment of the Hoshida S1 subtype60 and depletion of the S2 subtype among patients with MASH-HCC relative to HCC of mixed aetiology shown in Extended Data Fig. 1o.
Microarray
Microarray CEL files were downloaded from GSE164760. Normalization was performed using the robust multiarray average expression method implemented in Affy (v1.74.0). The HGU219 custom chip definition file was obtained from the Brainarray Microarray Lab (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/) and used for mapping probes to gene identifiers in a one-to-one format. Surrogate variable analysis was performed using sva (v3.44.0) to identify the presence of any unknown sources of variation by modelling different types of tissue as the variable of interest.
Differential gene expression analysis
For RNA-seq, overall and timepoint-specific differential gene expression results were obtained using the DESeq2 package (v1.36.0) by modelling the additive and interaction effect of timepoint and Acly KO. To obtain the overall result, we adjusted association of gene expression with Acly KO by a binary indicator of experimental timepoint (that is, early or late) as a confounding variable using an additive model (that is, time + Acly KO). Timepoint-specific results were obtained by modelling gene expression association using only samples belonging to either early or late timepoints. Finally, we modelled the modifying effect of timepoint on differential gene expression by using an interaction model (that is, time × Acly KO). We used a false discovery-adjusted threshold of 5% to determine gene significance. Individual sample read counts underwent variance stabilizing transformation. Gene ranks were obtained for downstream gene set enrichment analysis by performing natural logarithmic transformation of P value and applying the numerical sign of log2 fold change based on DESeq2 result.
Differential expression analysis for microarray data was performed using limma (v3.52.3). Comparisons between tissue were performed by specifying all combinations of contrast involving MASH-HCC. Comparison using ACLY expression level as an independent variable was performed by first using a linear model to identify overall fold change direction followed by a natural cubic splines model with 5 degrees of freedom to identify gene significance.
WGCNA
WGCNA (v1.71) in mouse tumour tissues was restricted to genes with greater than or equal to 15 counts in 75% of samples. A soft power of 14 was selected, resulting in a scale-free topology fit of 0.875 WGCNA for human MASH-HCC samples utilized a soft power of 12, resulting in a scale-free topology fit of 0.812. We constructed signed module networks for both mouse and human datasets using the robust correlation method and restricted the number of excluded outliers to 0.10 as recommended. Modules below the hierarchical clustering height of 0.25 were merged. Significant module genes and associated traits were identified using Pearson correlation and asymptotic P value for correlation.
Gene Ontology and gene set enrichment analysis
Over-representation and gene set enrichment analysis were conducted using clusterProfiler (v4.4.4). For over-representation analyses, we further calculated fold change as the ratio of a gene-to-background ratio. We used an adjusted P value of 0.05 as the threshold for significance. To identify clusters of similar Gene Ontology terms, we utilized the semantic similarity and binary cut method implemented in simplifyEnrichment (v1.6.1).
Cell-type deconvolution
Mouse cell-type deconvolution was performed using transcript per million normalized gene expression data and mMCP-counter (v1.1.0). Validation of B cell enrichment was performed using methods implemented in the online TIMER2.0 tool61. For human cell-type deconvolution, we used TIMER2.0 in addition to the ESTIMATE package (v1.0.13). We filtered cell types for those containing greater than 0 count in at least 75% of samples. Association with gene expression was assessed using univariate linear regression. Significance was determined using a false discovery-adjusted threshold of 5%.
Spatial transcriptomic analysis
Liver tissues were collected and fixed with 10% neutral-buffered formalin for 36–48 h. After fixation, the samples were immersed in a 70% alcohol solution. The liver tissues were then processed, paraffin-embedded and sectioned. RNA quality of formalin-fixed, paraffin-embedded tissues was tested by TapeStation system with DV200 > 30% (DV200 (distribution value 200) refers to the percentage of RNA fragments that are greater than 200 nucleotides in length). Slides with 5 µm of tissue sections were processed with deparaffinization, H&E staining, imaging, decrosslinking, hybridization, ligation, probe extension, pre-amplification and probe-based library construction to generate gene expression libraries for each tissue for sequencing. Images were taken using the Nikon 90i Eclipse upright microscope. RNA-seq was performed using the Illumina NextSeq 2000 (P2 Flow cell, 2 × 50 bp configuration) system. Sequence data from formalin-fixed, paraffin-embedded tissues were analysed using the Space Ranger count pipeline from 10X Genomics for raw data processing and quality control. Downstream analyses included quality control, normalization, dimensional reduction and clustering, variable gene selection, spatially variable feature detection, annotation, differential expression and integration with multiple samples using Seurat62. Spatial transcriptomics data analyses were performed using the Linux system, R, RStudio software and Python programming language. The B cell subtypes within the tumours were examined using established markers63.
scRNA-seq analysis
Frozen mouse liver samples (WD-DEN and WD-CCl4 HCC models) were minced, fixed and dissociated following the 10X Genomics ‘tissue fixation and dissociation for chromium-fixed RNA profiling’ protocol (document number CG000553, Rev B). Probe hybridization, library construction and sequencing followed the 10X Genomics ‘chromium-fixed RNA profiling reagent kits for multiplexed Samples’ (document number CG000527, Rev D). Sequencing was performed using the Illumina Novaseq 6000 sequencing system (S4, 2 × 150, 2–2.5 billion reads, targeting 5,000 cells per sample at a depth of 25,000 read pairs per cell). Demultiplexing was performed using Cell Ranger via the 10X Genomics Cloud Analysis platform. Demultiplexed samples were integrated by condition and processed in R using Seurat v5.
Mouse–human correlation analysis was performed in R. Mouse gene symbols were converted to their 1-to-1 orthologous human EnsemblIDs using the biomaRt package. Normalized count data were filtered for each dataset to include only genes common between all datasets. Each dataset was then filtered to include only cells expressing specific marker genes (those encoding CD45, AFP or GPC3, respectively). Pairwise Pearson correlation values were calculated between the human MASH-HCC data64 and WD-CCl4 mouse data15, using normalized gene expression in the relevant cell type. Results were visualized with the pheatmap package. Human gene-specific mean expression data were generated with Seurat (v5) in R. Wilcoxon rank-sum tests were used to assess the significance of differential expression of ACLY and SLC27A2 in different human conditions, relative to the MASH-HCC condition (P < 0.05 considered significant).
Single-nucleus sequencing analysis
Single-nucleus sequencing of cells derived from healthy, MASLD, MASH-HCC tumour and tumour-adjacent tissue were obtained from GSE174748 and GSE189175 (ref. 64) and processed using the Seurat package (v5.1.0) in R. We applied sctransform to normalize RNA read counts using the default selection of 3,000 genes as the number of variable features. Transformed counts underwent dimension reduction using principal component analysis. The 30 most variable components were then selected for uniform manifold approximation and projection. Clusters were then identified using the Louvain clustering method at a resolution of 0.2. Cell types were identified using the automated cell-type assignment pipeline as implemented in the sc-type package in R65. We utilized the custom gene set containing liver-specific cell-type expression markers provided by the GepLiver resource66. SCT normalized counts were used for this analysis. Low-confidence clusters based on the default score threshold was assigned with the ‘unknown’ cell type. The proportion of cells expressing ACLY was determined for hepatocytes and malignant hepatocytes and compared between tumour and non-tumour adjacent tissues using χ2 test of independence.
MIBI-TOF methods
Tissue staining
Tissues were sectioned (4-µm-thick sections) onto gold-coated MIBI slides. Slides were baked at 65 °C for 1 h, followed by deparaffinization and rehydration in sequential washes in xylene (3×), 100% ethanol (3×), 95% ethanol (2×), 70% ethanol (2×) and MIBI-water. Antigen retrieval in a pH 9 Target Antigen Retrieval Solution (DAKO Agilent) occurred at 125 °C for 40 min in a Decloaking Chamber (Biocare Medical). After cooling to room temperature, slides were washed twice in TBS-T (Ionpath). The tissue was incubated in a blocking buffer consisting of 3% normal donkey serum (Jackson ImmunoResearch) in TBS-T for 20 min. The antibody panel consisting of metal-tagged antibodies supplied by Ionpath was diluted in blocking buffer and added to the tissue for overnight incubation at 4 °C in a moisture chamber. After overnight incubation, tissues were fixed and dehydrated in sequential washes in TBS-T (3×), 2% glutaraldehyde (5 min), Tris pH 8.5 (3×), MIBI-water (2×), 70% ethanol (2×), 90% ethanol (2×), 95% ethanol (2×) and 100% ethanol (3×). Slides were stored in a desiccator before MIBIscope analysis. The antibody panels and reagents are listed in Supplementary Tables 3 and 4.
MIBI-TOF image acquisition
Spectral images of stained liver lesions were collected using an Ionpath MIBIscope with multiplexed ion-beam imaging technology. Xenon primary ions from a Hyperion ion gun rastered across the slide to sputter stained tissue, which was detected by mass spectrometry by time of flight to reconstruct the spectral images, on a pixel-by-pixel basis, of each channel consisting of a single stained antibody. A more detailed description of the multiplexed ion-beam imaging technology has been previously described42. Spectral images of 400 × 400-µm regions of interest (ROIs) were selected with the assistance of a pathologist and the images were collected using an Ionpath MIBIscope with multiplexed ion-beam imaging technology. These high-resolution images were denoised and filtered, and with the nuclear and membrane channels, we created segmentation masks using Mesmer67. For Acly-KO cohort, 35 400 × 400-µm regions for each experimental group, representing 43,974 cell objects, were acquired, whereas for the EVT0185 cohort, 24 400 × 400-µm regions for each experimental group were collected for a total of 30,191 cell objects.
Low-level image processing
Multiplexed raw image sets were denoised and aggregate filtered using Ionpath’s MIBI/O software using the default correction settings. Processed image data were stored as TIFF files for further data processing and analysis.
Cellular segmentation
Whole-cell segmentation was performed with the input of the nuclear stained and the membrane-stained marker channels using Mesmer67, and the segmentation mask images were stored as TIFF files for further analysis in R Studio.
Single-cell phenotyping
Single-cell data were extracted for all cell objects defined by the segmentation masks using a custom R script and packages described in Supplementary Table 5 (refs. 68,69,70,71). The aim of the single-cell analysis was to classify each of the objects in terms of their individual marker intensities to construct a profile of the tissue architecture and identify the proximity and relative spatial distribution of cells and features across the tumours. Marker intensities were asinh transformed by a cofactor of 1. To classify cell types based on their marker expression levels, the FlowSOM clustering algorithm was used with the Bioconductor ‘FlowSOM’ R package72. The algorithm clustered the single cells from the cohort into 100 FlowSOM clusters. By inspecting a heatmap displaying normalized individual marker intensities, we annotated each of the 100 clusters into ten cell types.
Selection of representative ROIs for cellular neighbourhood spatial analysis
The tumour ROIs were selected based on the presence or high abundance of tumour-infiltrating lymphocytes. Regions with the highest CD45-positive marker expression were selectively chosen as the representative of the cohort. The invasive margin was defined at the border of the malignant tumour and mostly consisted of approximately 50% malignant and 50% non-malignant tissue. The tumour and non-tumour regions were omitted from analysis. For each treatment group, four tumours were sampled for a total area of 1.6 mm2.
Cellular neighbourhood analysis
We used the imcRtools package to detect cellular neighbourhoods. A spatial graph was constructed by detecting the k-nearest neighbours in 2D per cell73. First, for each cell, we used the aggregateNeigbors function to compute the fraction of cells of a certain cell type among its neighbours. Second, for each cell, the function aggregated the expression counts across all neighbouring cells. On the basis of the fraction of the different cell types among the ten nearest neighbours, k-means clustering was used to group cells into a user-defined number of cellular neighbourhoods. The choice of six cellular neighbourhoods was determined by a parameter sweep across varying k values.
Lipid droplet analysis
Morphometric lipid droplet analysis was adapted from the previously published article74. In brief, H&E sections of tumour-bearing livers were processed using ImageJ for the average size of lipid droplets and average lipid area. Original H&E-stained slides were converted to an 8-bit greyscale image, which was then black and white inverted. These images were used for lipid droplet analysis using the following plugin: run (‘Analyze Particles…’, ‘size = 50–20,000’, circularity = 0.50–1.00 display summarize’), which defined the upper and lower limits to a lipid droplet area (50–20,000 µm2)75 to exclude irregular-shaped structures (lipid droplets are more spherical).
Tumour free fatty acids
Quantitation of free fatty acid levels was carried out using ultra-high-performance liquid chromatography–multiple reaction monitoring–mass spectrometry (UPLCMRM–MS) method as previously described76. In brief, a stock solution of fatty acids was prepared with their standard substances in LC–MS grade isopropanol:methanol (1:1). This solution was serially diluted 1:4 (v/v) to make 10 working standard solutions with the same solvent. A volume of the extractant equivalent to 2 mg of raw tissue was dried under a nitrogen gas flow. The residue was resuspended in 20 µl of LC–MS grade isopropanol:methanol (1:1). Each sample solution or 20 µl of each standard solution was then mixed in turn with 40 μl of 200 mM 3-nitro phenylhydrazine (3-NPH)-HCl solution and 40 μl of 150-mM EDC-HCl-6% pyridine solution. The mixtures were allowed to react at 40 °C for 60 min. After the reaction, 50 μl of each solution was mixed with 100 μl of a mixture of 13C6-3NPH derivatives of all the targeted organic acids. Of aliquots of the resultant solutions, 6 µl was injected onto a C18 column (2.1 × 100 mm, 2.5 µm) to run UPLCMRM–MS with (−) ion detection on an Agilent 1290 UHPLC system coupled to a Sciex 4000 QTRAP MS instrument, with the use of 1 mM ammonium acetate in water (A) and acetonitrile:isopropanol (1:1, v/v) as the mobile phase for binary-solvent gradient elution in a range of 30–100% B over 16 min at 0.4 ml min−1 and 60 °C.
Tumour TCA metabolites
The tumour tissues isolated were snap-frozen and stored in liquid nitrogen. Around 20–30 mg tissues from each group were chipped and homogenized (with 80% methanol)77. The supernatant was dried for gas chromatography (GC)–MS sample preparation78. Dried samples were further derivatized in 50 μl of 10 mg ml−1 methoxamine hydrochloride for 60 min at 42 °C, followed by 100 μl of N-tert-butyldimethylsilyl-N-methyl trifluoroacetamide for 90 min at 72 °C. Next, TCA metabolites were separated by Agilent 7890B gas chromatograph with HP-5ms Ultra Inert GC column (30 m, 0.25 mm, 0.25 µm, 7 inch; 19091S-433UI, Agilent). Metabolites were then analysed by Agilent 5977B mass-selective detector, using full-scan mode. Total metabolite abundances were measured as the area of the total ion counts normalized to protein content.
Tumour CXCL13 protein
Liver tumour CXCL13 protein levels were determined using the mouse CXCL13/BLC/BCA-1 Quantikine ELISA Kit (MCX130, R&D Systems) following the manufacturer’s instructions. In brief, 50 µl of tissue homogenate was added to each well of a 96-well plate coated with a monoclonal antibody specific for mouse BLC or BCA-1 for 2 h. Next, the plates were washed with wash buffer and 100 µl mouse BLC–BCA-1 conjugate was added to each well for the next 2 h followed by the addition of substrate and stop solution. The optical density was measured using a microplate reader set at 450 nm.
Immunofluorescence microscopy
Paraffin-embedded, 5-µm sections of tumour-bearing livers were used for immunofluorescence. Slides were deparaffinized, rehydrated and subjected to antigen retrieval (10 mM sodium citrate buffer). Slides were blocked in BSA and subsequently incubated with primary antibody overnight at a concentration of 1:500 CCP3 (Cell Signaling Technology) and Ki67 (Thermo Fisher). Fluorophore-conjugated secondary antibodies were then applied to tissues and mounted with coverslips. Slides were imaged on an inverted confocal microscope (Leica Microscope Systems). Representative images were acquired and analysed using ImageJ analysis.
Cell-free activity assays for ACLY, ACC1, ACC2, ACSS2 and AMPK
All cell-free assays were conducted at Reaction Biology. Human ACC1 (50202), ACC2 (50201) and ACLY (50255) were obtained from BPS Biosciences. Recombinant protein of human ACSS2, transcript variant 1 (accession no. NM_018677) tagged with MYC–DDK was obtained from Origene (TP304260). ADP-Glo (V9101, Promega) was used to measure ACLY, ACC1 and ACC2 enzyme activity following the manufacturer’s instructions. The assay was performed in two steps: (1) after the specific enzyme-mediated reaction that utilizes ATP and produces ADP, ADP-Glo reagent was added to terminate the kinase reaction and depleted the remaining ATP; and (2) kinase detection reagent was added to convert ADP to ATP and allow the newly synthesized ATP to be measured using a luciferase–luciferin reaction. The light generated, measured by the luminometer, correlated to the amount of ADP generated, which is indicative of ACLY, ACC1 or ACC2 activity. ACSS2 activity was assessed using the Bellbrook Transcreener AMP2/GMP2 Assay kit, a far-red competitive fluorescence polarization assay. AMP produced in the reaction was detected by AMP/GMP Alexa fluor 633 tracer bound to an AMP2/GMP2 antibody. AMP displaced the tracer allowing free rotation of the fluorophore and decreased fluorescence polarization, which was quantified using an appropriate multimode plate reader. Human AMPKα1–AMPKβ1–AMPKγ1 heterotrimers were co-incubated with the AMPK substrate peptide at 20 μM, AMP at 125 μM and ATP at 10 μM using the HotSpot assay.
ACLY competitive assay
Human recombinant ACLY was obtained from Sino Biological (11769-H07B). Recombinant full-length hACLY (accession no. P53396) was expressed in insect cells using baculovirus expression host, with a poly histidine tag at the N terminus. The molecular weight was 123 kDa. The ACLY ADP-Glo assays were performed at room temperature. Enzyme and sodium citrate were prepared in reaction buffer at three times the reaction concentration and 5 µl was added to Corning 3572 reaction plate wells. Of compound concentrations, 100× were prepared by serial dilution in 100% DMSO. Then, the compound was transferred to the reaction plate, which contains enzyme and sodium citrate, using acoustic technology. The compound and enzyme were pre-incubated for 15 min at room temperature. Then, 5 µl of 3× coenzyme A was delivered to the reaction well, followed by 5 µl of 3× ATP to start the reaction. The assay was incubated for 2 h at room temperature. For the background wells, all assay components were added without enzyme. After a 2-h reaction, ADP-Glo reagent was added to stop the reaction and incubated for 40 min at room temperature following detection kit protocol. Then, ADP-Glo detection was added and incubated for 30 min at room temperature following the detection kit protocol. The luminescence signal was measured after 30 min of incubation with ADP-Glo detection. The luminescence signal was then converted to µM ADP using ADP standard curve. The compound was tested in triplicate and each dataset was analysed separately.
Detection of EVT0185-CoA thioester using LC–MS/MS method
HEK293 cell culture and transfection validation
HEK293 cell line was obtained from American Type Culture Collection (ATCC) and grown in Dulbecco’s modified eagle medium (319-005-CL, Wisent Bioproducts) supplemented with 10% FBS (098150, Wisent Bioproducts) in a humidified incubator at 37 °C and 5% CO2. HEK293 cells were transfected with expression plasmid for SLC27A1 (NM_198580, human tagged ORF clone; RC209285, OriGene), SLC27A2 (NM_003645, human tagged ORF clone; RC221033, OriGene), SLC27A4 (NM_005094, human tagged ORF clone; RC209557, OriGene), SLC27A5 (NM_012254, human tagged ORF clone; RC215758, Origene) or control vector (pCMV6-entry mammalian expression vector; PS100001, OriGene). Transfected cell lines were supplemented with G418 (A1720, Sigma-Aldrich) during maintenance at a concentration of 800 µg ml−1. Transfected SLC27A2 protein was detected by western blot using the antibody DYK tag (2368, Cell Signaling); while endogenous SLC27A2 was detected using the polyclonal antibody (PA5-30420, Invitrogen).
Cell treatment and extraction
The day before the experiment, HEK293 cells overexpressing SLC27A1/A2/A4/A5 were plated in six-well plates at 1–1.5 × 105 cells per cm2 in Dulbecco’s modified eagle medium and cultured overnight. Cells were treated with 30 µM of EVT0185 for 3 h. At the end of the treatment period, medium was removed, cells were washed once with ice-cold PBS and were flash frozen in liquid N2. For extraction, plates were placed on ice, and cells were scraped in 266 µl of PBS per well. The cell suspension from three wells were pooled (approximately 800 µl total volume) and added to 1 ml of ice-cold 2:1 dichloromethane:methanol in a glass vial. The samples were vortexed for 2 × 12 s and then centrifuged at 300g for 5 min at 4 °C. The upper aqueous phase was transferred to a new tube and filtered through a 0.45-µm PTFE filter. Samples were delivered to the Centre for Microbial Chemical Biology at McMaster University for LC–MS/MS analysis.
LS–MS/MS analysis
Samples were evaporated using a Turbovap LV (Biotage) evaporation system with the water bath set to 30 °C. Dried samples were resuspended in 100 µl of methanol, then analysed by injecting 5 µl into an Agilent 1290 Infinity series HPLC (Agilent) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Separation of the analytes was achieved on an Agilent Eclipse XDB-C18 (100 mm × 2.1 mm, internal diameter (i.d.), 3.5 µm) analytical column using mobile phases consisting of (A) 10 mM ammonium acetate in water, pH 8.5, and (B) acetonitrile and a constant flow of 0.2 ml min−1. The analytical column was maintained at 30 °C. The separation of analytes was achieved using a gradient starting at 20% B, which was held for 1.5 min. The gradient increased to 95% B over the next 3.5 min and was held for 4.5 min. The column was then re-equilibrated back to the initial conditions over the final 5.5 min. The total run time was 16 min. The HPLC eluent was introduced into the mass spectrometer using electrospray ionization in positive mode with a spray voltage of 3.6 kV. The mass spectrometer was set to acquire spectra in data-dependent acquisition mode. The full MS scan was set to an m/z range of 100–2,000 in the ion trap with a resolution of 30,000. The MS/MS was performed in the Orbitrap with a collision induced dissociation (CID) collision energy of 35. The activation time was set at 30 ms with the activation parameter q = 0.250 and an isolation window of 1.5 m/z. The resulting mass spectra were analysed using the Xcalibur 2.1 Qualitative software package.
Clonogenic assay
Hep3B (HB-8064) human hepatocarcinoma and Hepa1-6 (CRL-1830) mouse hepatoma cell lines were purchased from the ATCC and grown in eagle’s minimum essential medium (10-009-CV, Mediatech) and Dulbecco’s modified eagle medium (319-005-CL, Wisent Bioproducts), respectively, supplemented with 10% FBS (098150, Wisent Bioproducts) and 1% penicillin–streptomycin (15140122, Gibco, Thermo Fisher Scientific) in a humidified incubator at 5% CO2/95% air. Bempedoic acid was purchased from MedChemExpress. Hep3B and Hepa1-6 cells were seeded in 12-well plates at a cell density of 1,000 and 500 cells per well, respectively. On day 2, media in each well were aspirated and replaced with 1,000 µl of fresh complete media and cells were treated with or without the respective drugs in an increasing concentration followed by the incubation of the cells for 7 days in an incubator. On day 9, the media in each well were aspirated and cells were fixed with 10% formalin (500 µl) for 10 min at room temperature. Formalin was aspirated and the plates were washed with 1× PBS and stained with crystal violet for another 10 min. After 10 min of staining, crystal violet was poured over the cells, rinsed with tap water for three times and the plates were dried overnight. The next day, colonies (more than 50 cells) were counted and analysed.
Cryo-EM studies on ACLY–EVT0185-CoA
The pTrcHis2-hACLY expression construct35 for full-length human ACLY (Uniprot ID P53396-2) in frame with C-terminal MYC and His tags was transformed in BL21(DE3) cells. Cultures were grown in Luria–Bertani medium at 28 °C and expression was induced by the addition of 1 mM IPTG at an optical density at 600 nm of 0.6–0.7. After overnight expression, the bacterial culture was collected by centrifugation and bacterial cells were resuspended in IMAC-binding buffer, consisting of 50 mM sodium phosphate, pH 7.4, and 150 mM NaCl supplemented with cOmplete protease inhibitor cocktail without EDTA (Roche). Bacterial cells were lysed by sonication and insoluble material was removed by centrifugation. The resulting supernatant was clarified using a 0.22-μm filter and loaded onto a Ni Sepharose column equilibrated with IMAC-binding buffer. The IMAC column was washed and His-tagged ACLY was eluted with binding buffer supplemented with increasing concentrations of imidazole. Elution fractions were pooled and concentrated using ultracentrifugation. ACLY was further purified by size-exclusion chromatography using HiLoad 16/600 Superdex 200 and Superose 6 (Increase) columns with 20 mM HEPES, pH 7.4, and 150 mM NaCl as a running buffer. The top fractions from the final size-exclusion chromatography elution peak were pooled and concentrated to 10 mg ml−1, aliquoted, flash frozen and stored at −80 °C in a freezer until further use.
For cryo-EM grid preparation, purified ACLY (10 mg ml−1 in HBS buffer) was supplemented with 0.5% CHAPSO, 1 mM Mg2ATP and 4 mM EVT0185-CoA (freshly prepared from powder). Of the sample, 4 µl was applied to a glow-discharged C-Flat 1.2/1.3 300 mesh copper grid (Protochips), blotted for 4.5 s under 99% humidity at 22 °C and plunged into liquid ethane using a GP2 Leica grid plunger. Grids were screened using a JEOL 1400 Plus microscope equipped with a JEOL Ruby CCD camera at the VIB Bioimaging Core Ghent. Data collection was performed using a JEOL cryoARM 300 microscope equipped with a 6k × 4k GATAN K3 detector resulting in 7,263 movies with a raw pixel size of 0.72 Å (BECM). Movies were processed via patch-based motion correction and contrast transfer function estimation as implemented in cryoSPARC (v3.1.0)79.
Particles were extracted with a box size of 480 pixels with 2× binning. Good-quality 2D classes were initially obtained via the Blob Picker job in cryoSPARC, followed by template-based picking and neural network-based particle picking via TOPAZ as implemented in cryoSPARC80. Ensuing 2D classification, 2D class selection and removal of potential duplicate particles within a distance of 150 Å resulted in a particle set of 210,900 particles. Ab initio 3D classification (number of classes = 5) followed by homogeneous and non-uniform refinement81 without application of symmetry resulted in a 3D reconstruction that had a resolution of 3.7 Å following a gold-standard non-uniform refinement and represented the pseudo-D2-symmetric ACLY tetramer. In this cryo-EM map, the central, tetrameric CSH module was markedly better defined than the N-terminal CCS modules, indicating structural heterogeneity of the CCS modules with respect to the central CSH module.
To address this flexibility and possibly resolve the CCS–CSH assembly at a higher resolution, we applied symmetry expansion in combination with local refinement followed by 3D classification. Following homogeneous and non-uniform refinement in D2 symmetry, the associated particle set was re-extracted without binning (0.72 Å per pixel) and symmetry expanded around the D2 axes. Using the molmap function in Chimera, a volume blurred to 25 Å that contained one CCS arm and the central CSH module was generated and transformed into a mask with the Volume Tools job in cryoSPARC (map threshold = 0.09, dilation radius = 7.2 Å and soft padding width = 14.4 Å). Gold-standard local refinement was performed by limiting the rotation and shift search extent around the original consensus refinement poses to 20° and 10 Å, respectively, in combination with a Gaussian prior over the pose or shift magnitudes (with SDROT = 15 Å and SDSHIFT = 7 Å). The centre of mass of the mask was used as a fulcrum point. Ensuing 3D classification without alignment in cryoSPARC using the 3D classification job followed by another round of gold-standard local refinement resulted in a cryo-EM volume with a FSC0.143 resolution of 3.3 Å in which the atomic models for the CSS and CSH modules (extracted from Protein Data Bank (PDB) ID 6XHX) were fitted using Chimera and real-space refined in Phenix82 using reference restraints to the starting model. Cryo-EM map regions representing ligand density in the ATP-grasp fold domain of the CCS module and in the CoA-binding domain were of the CSH module modelled as Mg.ADP and the adenosine 3′-phosphate 5′-diphosphate moiety of bound EVT0185-CoA, respectively. Restraints for EVT0185-CoA were generated via the de Grade Web Server (https://grade.globalphasing.org)83.
A cryo-EM micrograph for the ACLY–EVT0185-CoA complex is provided in Extended Data Fig. 4b. Cryo-EM data and refinement statistics are summarized in Supplementary Table 2. Reported resolutions are based on the gold-standard FSC0.143 criterion84, and FSC curves were corrected for the effects of soft masking by high-resolution noise substitution85. A map-to-model correlation was calculated using phenix.mtriage using the independent half maps as input. Local resolution maps were computed using the blocres algorithm86 as implemented in cryoSPARC with an FSC threshold of 0.5. All representations of cryo-EM Coulomb potential density maps and structural models were prepared with ChimeraX87 and PyMol88. Cryo-EM map contour sigma levels have been reported based on map normalization in Coot.
Materials availability
The protein expression construct for full-length human ACLY is available via the BCCM/GeneCorner Plasmid Collection (http://bccm.belspo.be) through the following accession code: LMBP 11277 (pTrcHis2-hACLY).
Cryo-EM maps following global and local refinement and the real-space-refined model for the CCS–CSH assembly have been deposited in the Electron Microscopy Data Bank (EMDB) with the accession code EMD-53847 and in the PDB with the ID 9R90 (Extended PDB ID pdb_00009R90).
Generation of stable ACLY knockdown cell line
Cell culture
Hep3B cells (ATCC HB-8064) were cultured adherently using Eagle’s minimum essential medium (ATCC 30-2003) supplemented with 10% (v/v) FBS (10437-028, Gibco) and 1% (v/v) antibiotic–antimycotic solution (15240112, Gibco). HEK293T (ATCC CRL-3216) was cultured adherently using Dulbecco’s modified eagle medium, high glucose (11965092, Gibco) supplemented with 10% (v/v) FBS and 1% (v/v) antibiotic–antimycotic solution as aforementioned along with 1% (v/v) l-glutamine (25030081, Gibco), 1% (v/v) MEM non-essential amino acids solution (11140050, Gibco) and 1% (v/v) sodium pyruvate (11360070, Gibco).
Plasmid construction
Dox-inducible shACLY plasmid vectors were generated using the inducible EZ-Tet-pLKO-Hygro vector (Addgene plasmid no. 85972). Two ACLY shRNA sequences—5′-CGTGAGAGCAATTCGAGATTA and 5′-GGCATGTCCAAGCTCAA—and one non-targeting shRNA sequence (5′-CCTAAGGTTAAGTCGCCCTCG) were ligated, transformed, amplified and validated using established protocol as previously described89.
Lentivirus production
A second generation lentivirus production system was used to generate high-titre lentivirus. HEK293T cells were incubated with psPAX2 (Addgene plasmid no. 12260), pMD2.g (Addgene plasmid no. 12259) and the EZ-Tet-pLKO-Hygro (Addgene plasmid no. 85972) transfer plasmid at a ratio of 1:1:2 pmol in the presence of Lipofectamine 2000 in Opti-MEM. Sixteen hours post-transfection, media were changed to 1% bovine serum albumin (A1470, Sigma-Aldrich) and 1 mM Na-butyrate (3850, Tocris) supplemented with antibiotic-free DMEM (11965092, Gibco) for HEK293T cell culture. Forty-eight hours post-transfection, lentivirus media were collected and mixed with Lenti-X concentrator (631232, Takara) before storing overnight at 4 °C. Next day, lentivirus was concentrated through centrifugation at 1,500g for 45 min at 4 °C. The resulting pellet was resuspended at 1:10 of initial media volume, aliquoted and frozen in liquid nitrogen for storage.
Lentivirus transduction
An initial dilution of 1:50 v/v followed by serial dilutions of 1:4 v/v were used to transduce target cell lines at 60,000 cells per well in six-well plates using 5 µg ml−1 polybrene (107689, Sigma-Aldrich) supplemented cell culture media. Cells were transduced for 24 h, followed by recovery in lentivirus-free media for another 24 h before selection of successfully transduced cells using a pre-determined, cell line-dependent concentration of hygromycin B (H3274, Sigma-Aldrich) or puromycin (P8833, Sigma-Aldrich) for 7 days. Surviving cells post-antibiotic selection were further propagated to establish stable cell line.
DNL assay
Cells were incubated in media supplemented with 1 µCi [14C] glucose (NEC042V250UC, Perkin Elmer) and [3H] acetate (NET003005MC, Perkin Elmer) for 4 h. Subsequently, cells were washed and scraped in PBS (10010023, Gibco). Lipids were extracted using a chloroform:methanol (1:2) solution, vortexed for 30 s and centrifuged at 13,000g for 10 min to isolate cellular debris and protein fractions from lipid supernatant. Lipids were further purified with chloroform:water (1:1) solution via vortex and centrifugation as described. Peptide concentration was quantified using the Pierce BCA Protein Assay Kit (23225, Thermo Scientific) for normalization calculation. Of lipid supernatant, 100 µl was extracted from the non-polar chloroform phase and mixed with 5 ml of Ultima Gold Scintillation Fluid (6013329, Perkin Elmer) for radioactivity quantification.
Fatty acid oxidation assay
Cells were incubated with serum-free cell culture media supplemented with 2% bovine serum albumin (A1470, Sigma-Aldrich), 500 µM sodium palmitate (P9787, Sigma-Aldrich), 0.5 µCi ml−1 [14C] palmitic acid (NEC075H050UC, Perkin Elmer) and 1 mM l-carnitine (11242008001, Roche) for 4 h. Cell culture supernatant was collected at the end of incubation and mixed with 1 ml of 1 M acetic acid (34256-1L-R, Fluka). Solution was sealed in a glass vial and shaken at 75 rpm at room temperature for 2 h. Released CO2 was absorbed in 450 µl of 1 M benzethonium hydroxide (B2156, Sigma-Aldrich) during this time. Subsequently, tube containing benzethonium hydroxide was mixed with 5 ml of Ultima Gold Scintillation Fluid (6013329, Perkin Elmer) for radioactivity quantification.
For western blotting, cells were washed with cold PBS (10010023, Gibco) and scraped with 100–200 µl of cold cell lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 100 mM NaF, 10 mM Na-pyrophosphate, 5 mM EDTA, 250 mM sucrose, 1 mM dithiothreitol and 1 mM Na-orthovanadate, 1% Triton X-100 and cOmplete protease inhibitor cocktail; 11836153001, Roche). Solution samples were collected and centrifuged at 13,000g for 10 min at 4 °C to isolate protein fraction from cell debris. Protein samples were quantified for peptide concentration using the Pierce BCA Protein Assay Kit (23225, Thermo Scientific) according to instruction. Samples were then diluted to 1 µg µl−1 using 4× SDS sample buffer (40% glycerol, 240 mM, Tris-HCl, pH 6.8, 8% SDS, 0.04% bromophenol blue, 5% β-mercaptoethanol and 20 mM dithiothreitol) and cell lysis buffer. Samples were then heated for 5 min at 95 °C before immunoblotting. SDS polyacrylamide gels were prepared at 10% or 12% polyacrylamide concentration dependent on protein size. Of the protein sample, 25 µg was loaded into each lane. Gel electrophoresis was performed first at 90 V as protein travels through the stacking gel and then at 120 V, through the separating gel. Protein samples were then transferred onto a PVDF membrane (03010040001, Roche) at 4 °C for 90 min following the standard wet transfer protocol. Subsequently, membranes were rinsed and rocked gently with 1× TBS (50 mM Tris, 150 mM NaCl and 1 M HCl, pH 7.4) for three cycles of 5 min each then blocked using a TBST-BSA solution (1× TBS, 0.5% bovine serum albumin and 0.1% Tween-20) for 1 h at room temperature. Primary antibodies (ACLY (13390; 1:1,000 dilution); p-ACLY S455 (4331; 1:1,000 dilution); and β-actin HRP conjugate (12620; 1:5,000 dilution); all Cell Signaling Technology) were diluted at 1:1,000 in TBST-BSA solution. Membranes were then cut according to protein sizes and incubated in respective antibody solutions overnight at 4 °C with gentle rocking. The next day, primary antibody solutions were removed, and membranes were washed using TBST. Subsequently, membranes were incubated at room temperature with gentle rocking for 1 h in solutions containing 1:10,000 dilution of species-specific secondary antibody conjugated to HRP (anti-rabbit IgG HRP linked; 7074, Cell Signaling Technology). At the conclusion of incubation, membranes were washed again in TBST solution and kept in TBS solution until imaging and for storage.
Human PBMC culture and T lymphocyte and B lymphocyte proliferation assay
Peripheral blood mononuclear cells (PBMCs) were isolated from donors using the Ficoll Paque density gradient method. PBMCs were then washed with RPMI1640 and pelleted. Cells were resuspended with 10 ml of 3.5 µM CellTrace CFSE (C34570, Thermo Fisher) and incubated at room temperature for 20 min. After incubation, 40 ml complete RPMI1640 was added for the next 5 min to absorb excess CFSE. PBMCs were then washed, resuspended in complete RPMI1640 (10% FBS, 1% penicillin–streptomycin and 1% l-glutamine) and plated at a cell density of 1 × 106 cells per well in 12-well plates.
For T cell proliferation, PBMCs were induced with 3 µg ml−1 of concanavalin A (C5275, Sigma-Aldrich) and 10 ng ml−1 of human recombinant IL-2 (200-02, Preprotech). For B cell proliferation, PBMCs were induced with 1 µg ml−1 of R848 (36611, mAb Tech) and 10 ng ml−1 of IL-2 (36611, mAb Tech). EVT0185 was diluted in DMSO and was added at the final concentration of 0.1 µM, 0.3 µM, 1 µM, 3 µM and 10 µM. Next, PBMCs were incubated at 37 °C and 5% CO2 incubator for 5 days. Cells were then harvested, stained with E780 Live/Dead stain (C34570, Thermo Fisher), CD3 (300434, BioLegend) and CD19 (302241, BioLegend) antibody and analysed using flow cytometry. The results were further analysed using the FlowJo software. After 5 days, cells were then prepared for flow cytometry.
Flow cytometry
For the B cell depletion study, the tail vein blood was collected from each animal after 15 days of single injection with 250 μg of anti-CD20 (152104, BioLegend) or isotype (400566, BioLegend) antibody. The red blood cells were lysed twice with 1× red blood cell lysis buffer. The samples were then centrifuged at 1,500 rpm for 5 min at 4 °C. The cells were washed and blocked with Fc block (1:200; 553142, BD Biosciences) and stained with CD45.2 BV510 (1:25; 109838, BioLegend), B220 (1:100; 563894, BD Biosciences), CD19 (1:100; 152409, BioLegend) and 7AAD (1:100; A1310, Thermo Fisher Scientific). CytoFlex Flow Cytometer (Beckman Coulter Life Sciences) was used for data acquisition and was further analysed using FlowJo (v10.5).
For PBMC analysis, the cells were harvested after 5 days of treatment and stained with E780 Live/Dead stain (C34570, Thermo Fisher), CD3 (300434, BioLegend) and CD19 (302241, BioLegend) antibody and analysed using flow cytometry. The results were further analysed using the FlowJo software.
Statistical analysis and reproducibility
Independent t-test and analysis of variance (ANOVA) were performed using GraphPad Prism software (v9.5.1). Correlation and regression analysis were performed using R studio. All values are reported as mean ± s.e.m. unless stated otherwise. Differences were considered significant (∗) when P < 0.05. Statistical significance between two independent groups was analysed by unpaired t-test. Representative images shown in Figs. 1e and 5g and Extended Data Figs. 1n and 9d,e were repeated at least four times. Extended Data Fig. 3d,e was repeated two times. Extended Data Fig. 4b shows a representative cryo-EM micrograph from the dataset (7,263 micrographs in total), illustrating the particle distribution and image quality. Histological scores were analysed using the non-parametric Mann–Whitney test when comparing two independent groups. Wilcoxon’s test was used to analyse cell type and regional prevalence in MIBI-TOF images. Descriptions of additional software and statistical analysis used for transcriptomic analysis are mentioned in their respective methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.