Ethics information
All animal procedures were conducted in compliance with national and international ethical guidelines and regulations. Mouse experiments were approved by the local animal welfare authorities at Heidelberg University Interfaculty Biomedical Research Facility (T-64/17). Chicken experiments were conducted under UK Home Office licence PP9565661 and approved by the Roslin Institute Animal Welfare and Ethical Review Board Committee and the Linköping Council for Ethical Licensing of Animal Experiments (288-2019). Mice (Mus musculus; strain CD-1; RjOrl:SWISS; RRID: MGI:5603077) were purchased from Janvier Labs and euthanized by means of cervical dislocation. All chicken (Hy-Line Brown; Gallus gallus) management, maintenance and embryo manipulation followed the relevant regulatory guidelines.
Isolating, sexing and culturing PGCs
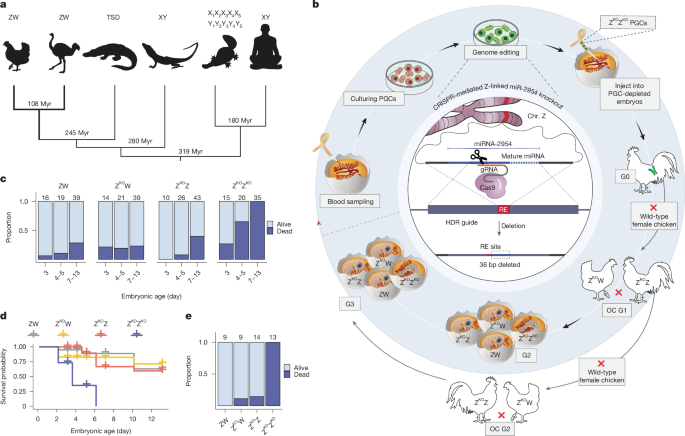
Genome editing in chickens involves the derivation and culturing of PGCs, performing genome editing on these cells and the subsequent injection of the edited cells into surrogate hosts depleted of their native PGCs21. Following the injection of the genetically edited PGCs into the gonads of sterile surrogate hosts, the resulting offspring will inherit the genetic modifications introduced into the PGCs21 (Fig. 1b and Extended Data Fig. 1). To establish miR-2954 KO lines, ten PGC lines were derived from the blood of Hy-Line Brown chicken embryos at Hamburger–Hamilton stage 16 (E2.5) and cultured according to previously described methods21. The sex of the PGC lines was determined according to previous studies21,54 on the basis of two sets of primers for one W-chromosome-specific gene and one autosomal gene (the control), respectively; the latter serves as a control for polymerase chain reaction (PCR) success (Supplementary Table 1). We cultured four male PGC lines and subsequently randomly selected one line for the KO experiment. This PGC line was cultured for 22 days in total before transfection.
Design of sgRNA and homology-directed repair template
Inducing double-stranded breaks at specific genomic loci, followed by homology-directed repair using a template, introduces precise nucleotide substitutions20. Using CHOPCHOP v.2 (ref. 55), we designed and tested five custom sgRNAs (Supplementary Table 1) to target the miR-2954 (MIR2954) locus (Gene ID: 100498678), located within the second intron of the DNA damage recognition and repair factor gene, XPA (ENSGALG00010009534), on the forward strand of chromosome Z (location: NC_052572.1: 71305174-71305241; reference genome: bGalGal1.mat.broiler.GRCg7b (GCF_016699485.2)). Additionally, we designed one single-stranded DNA oligonucleotide (ssODN) sequence as a repair template to exploit the homology-directed repair pathway. The ssODN repair template consisted of Ultramer DNA Oligonucleotides, custom-synthesized by Integrated DNA Technologies. The ssODN template contained homology arms flanking miR-2954, designed specifically to introduce a 36-bp deletion encompassing the entire mature miR-2954 sequence and part of its flanking pre-miRNA sequence. Additionally, we incorporated an EcoRI restriction endonuclease site (5′-GAATTC-3′) into this ssODN (Supplementary Table 1). These modifications effectively knock out miR-2954 and allow PCR-based genotyping for successful deletion events in both PGCs and the derived chickens (Extended Data Fig. 1 and Supplementary Table 1).
Genotyping
We designed PCR primers to amplify a 550-bp region within the second intron of the XPA gene, encompassing the targeted deletion site (Supplementary Table 1) using Primer-BLAST56. EcoRI restriction endonuclease enzyme specifically recognizes and cuts DNA at the restriction site (5′-GAATTC-3′). Following EcoRI digestion of this PCR product and subsequent gel electrophoresis, we expected to observe a single 550-bp band in wild-type individuals (ZZ and ZW) owing to the absence of the EcoRI restriction site, three bands (550, 298 and 221 bp) in heterozygote KO individuals (ZKOZ) owing to digestion of half of the product and two bands (298 and 221 bp) in homozygote males (ZKOZKO) and hemizygote females (ZKOW) owing to complete EcoRI restriction site digestion. This differential PCR band pattern served as a molecular signature for genotyping the individuals. PCR was performed using Phusion High-Fidelity PCR Master Mix with GC Buffer from New England Biolabs, in accordance with the manufacturer’s guidelines. The reaction mixture was prepared using 1.25 µl of 10 µM forward primer, 1.25 µl of 10 µM reverse primer, 0.75-µl dimethyl sulfoxide, 12.5 µl of 2X Phusion Master Mix and approximately 100 ng of DNA in 1 µl of water. The thermal cycling conditions were set as follows: an initial denaturation at 98 °C for 60 s, followed by 35 cycles of 98 °C for 10 s, 62 °C for 20 s and 72 °C for 20 s, concluding with a final extension at 72 °C for 10 min. To perform genotyping, we first extracted DNA from approximately 10,000 PGCs or embryonic tissues using DNeasy Blood & Tissue Kits from QIAGEN, according to the manufacturer’s protocol. We then conducted PCRs as described above and subjected the PCR products to EcoRI digestion using EcoRI-HF and rCutSmart buffer from New England Biolabs, following the manufacturer’s guidelines. Each reaction consisted of 5 µl of the PCR product, 1 µl of EcoRI-HF, 1 µl of rCutSmart buffer and 8 µl of water. The reactions were incubated at 37 °C for 30 min, followed by a 5-min heat inactivation at 65 °C. Alternatively, the genotypes of several samples were analysed on the basis of the size of the undigested PCR products using the Agilent Fragment Analyzer system. In this approach, a 550-bp band represented ZZ and ZW, a 520-bp band represented ZKOZKO and ZKOW and two bands (550 and 520 bp) in ZKOZ.
PGC transfection, selection and clonal expansion
We used a high-fidelity Cas9 variant (SpCas9-HF1), which significantly reduces off-target effects compared to wild-type Cas9 (ref. 19). For the expression of SpCas9-HF1 and sgRNAs in PGCs, we used the HF-PX459 (V2) expression vector, which also bears puromycin resistance as an antibiotic selection gene17 (Addgene plasmid 118632). We cloned all five sgRNAs individually into the plasmids according to previous descriptions17,20 and then tested the effectiveness of three of these plasmids harbouring sgRNAs 1–3. We transfected 1.5 µg of the vector and 0.5 µg of ssODNs into approximately 100,000 Hy-Line Brown PGCs using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). After 24 h in culture, the cells were treated with 0.6 µg ml−1 of puromycin for 48 h for the selection of successfully transfected cells. We cultured these cells for around 2 weeks and then genotyped them for the presence of deletions through EcoRI digestion of the PCR product. Using gRNA3, we observed a strong PCR band at 550 bp and two faint bands at approximately 300 and 220 bp. This pattern suggested the incorporation of the ssODN template in a subset of transfected PGCs. Accordingly, these PGCs were sorted using the BD FACSAria III Cell Sorter (BD Biosciences) into a 96-well plate at a rate of one cell per well to identify the clonal populations with the deletion of miR-2954. After 3 weeks of culturing, we screened the genotypes of 42 clonal PGC populations that survived and propagated. We identified four ZKOZ and two ZKOZKO clonal populations among them (6 of 42 clones were targeted). Subsequently, we cryopreserved the homozygote and heterozygote populations following established protocols21 and used one of the ZKOZKO populations for confirmation of the deletion and injection to surrogate hosts to generate the KO animals. To confirm the deletion of miR-2954, we performed PCR on the DNA obtained from the PGC line before transfection. The selected clonal ZKOZKO PGC population and the resulting PCR products were sequenced by Eurofins Genomics using their Sanger sequencing services (TubeSeq Service). Analysis of the sequences confirmed the deletion of miR-2954 and integration of the EcoRI site in accordance with the design of the provided ssODN repair template (Extended Data Fig. 1).
Generation of the G0 rooster
ZKOZKO PGCs were injected into surrogate host embryos using our previously described method21. In brief, we thawed the cryopreserved clonal ZKOZKO PGCs 7 days before the intended injection date and propagated them to a density of approximately 150,000 cells per well in a 24-well tissue culture plate. These cultured PGCs were pelleted by means of standard centrifugation and then resuspended in the PGC culture medium to achieve a concentration of 5,000 cells per microlitre. To this suspension, we added 0.1 μl of the chemical compound AP20187 (B/B) (25 mM) per 5 μl of PGC suspension. Approximately 1 μl of this mixture was aspirated into a microcapillary injection tube and injected into each iCaspase9 sterile embryo16 at Hamburger–Hamilton stages 15 and 16. AP20187 (B/B), present in the injected PGC mixture, induces the dimerization of the FK506-binding protein, leading to the activation of the attached caspase-9 protein and the induced apoptotic cell death of the endogenous PGCs in the iCaspase9 sterile embryos, thereby allowing the colonization of gonads by the injected ZKOZKO PGCs16. Injecting the clonal PGCs into 20 iCaspase9 sterile embryos resulted in hatching of 7 G0 chicks comprising 1 male and 6 females.
Generation of miR-2954 KO chickens
We maintained the male G0 and raised it to sexual maturity. This G0 was then paired with six Hy-Line Brown hens (same breed), producing ZKOZ and ZKOW individuals (OC G1). We then raised five male and six female OC G1 individuals to sexual maturity. One of these males was mated with the OC G1 females to generate second-generation (G2) embryos (ZKOZ, ZKOZKO, ZW or ZKOW) that were used for viability studies and tissue collection for gene expression analyses. A second OC G1 male, not involved in generating G2 individuals, was mated with six Hy-Line Brown females. This pairing produced OC G2 individuals for the genotypes ZZ, ZKOZ, ZW and ZKOW. Finally, upon reaching sexual maturity, a OC G2 ZKOZ rooster was mated with six OC G2 ZKOW hens to produce G3 embryos (ZKOZ, ZKOZKO, ZW or ZKOW). These G3 embryos were then used to confirm the phenotypes observed in the G2 generation (Fig. 1a).
Selection and processing of chicken embryos and tissues for RNA-seq analysis
Upon completing the genotyping and sexing of G2 embryos, we selected 36 embryos for RNA-seq. This selection included 18 E2 embryos (9 males and 9 females) (Hamburger–Hamilton stage 12), 9 E3 males (Hamburger–Hamilton stages 18 and 19) and 9 E5 males (Hamburger–Hamilton stages 24 and 25). For the E2 cohort, RNA extraction was performed on whole embryos after the removal of extra-embryonic membranes. This cohort included nine female embryos of various genotypes (three ZW, three ZKOW and three pure Hy-Line Brown ZW embryos (female embryos from the original stock), as a control for maternal effects on gene expression), and nine male embryos (three ZKOZ, three ZKOZKO and three ZZ genotypes). Given the low expression of miR-2954 in females and their survival, we then focused on gene expression in males. For the E3 and E5 cohorts, we investigated tissue-specific gene expression by dissecting the head, heart and rest of the body (referred to as the body) from each male embryo under a stereomicroscope, with all dissections performed in ice-cold PBS. Each tissue type from each embryo was represented by three replicates derived from three individuals. We note that all ZZ are pure Hy-Line Brown, and all other genotypes (ZW, ZKOW, ZKOZ and ZKOZKO) are G2.
RNA extraction and sequencing
A total of 72 samples from E2, E3 and E5 embryos were used for the generation of RNA-seq libraries. We extracted total RNA from whole embryos or dissected tissues using the AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN), following the manufacturer’s protocols. The RNA quality was assessed using the Fragment Analyzer system (Agilent), and all RNA quality numbers were equal to 10, indicating a lack of degradation. The RNA-seq libraries were prepared from 400 ng of RNA per sample using the NEBNext Ultra II RNA Library Prep Kit for Illumina sequencing on an Illumina NextSeq 2000 system, using NextSeq 2000 P3 Reagents (100 cycles), with samples multiplexed in two sets of 36.
Additionally, we generated small RNA libraries using RNA derived from the same E5 male samples (which were also used to generate RNA-seq libraries). This included the generation of small RNA libraries for RNA derived from ZZ (two replicates), ZKOZ (three replicates) and ZKOZKO (three replicates) for each tissue type (head, body and heart, respectively). These libraries were prepared using the NEBNext Small RNA Library Prep Set for Illumina and were sequenced on an Illumina NextSeq 550 system using NextSeq 500/550 High Output Kit v.2.5 (75 cycles), with samples multiplexed in two sets of 12.
Estimation of gene expression levels
The chicken reference genome (bGalGal1.mat.broiler.GRCg7b; GCA_016699485.1) and corresponding gene transfer format (GTF) annotation file were obtained from Ensembl57 (release 109). Raw reads from each library were aligned to the reference genome using STAR aligner v.2.7.2b (ref. 58). This alignment process involved generating STAR indices, aligning reads to the reference genome in an annotation-aware manner and quantifying the number of reads mapped to each gene using the quantMode GeneCounts option in STAR. The median uniquely mapped reads number across all samples was 34,703,339. The resulting gene count matrices, along with a metadata file containing sample information and the GTF file, were used to create a RangedSummarizedExperiment object. This object was imported into DESeq2 v.1.24.0 (ref. 58) for downstream analysis. Gene expression data were normalized using variance-stabilizing transformation (VST) through the vsn package v.3.52.0 in R v.4.1 (ref. 59) implemented in the DESeq2 package. Subsequently, principal component analysis (PCA) was conducted as implemented in the DESeq2 package to examine sample relationships and identify potential outliers. The PCA results revealed a clear clustering of samples (including biological replicates) for the respective tissues and ages without outliers, supporting the high quality of the expression data (Extended Data Fig. 3a).
Raw short RNA-seq data were preprocessed using a custom Bash script. Adaptor sequences were trimmed and reads were size-selected using Cutadapt v.4.4. The parameters set a maximum error rate of 0.25, targeted the adaptor sequence AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC with a minimum overlap of 6 nucleotides and allowed no indels while selecting for read lengths between 19 and 26 nucleotides. After trimming and size selection, the reads were aligned to the chicken reference genome using STAR following the ENCODE miRNA-seq pipeline60 (www.encodeproject.org/microrna/microrna-seq-encode4/) (May 2017). This alignment process included mapping to the miRNA subset of the chicken GTF gene annotation and quantifying the number of aligned reads in STAR. The median of the number of uniquely mapped reads across all samples was 432,069.
MiRNA target prediction in chicken, zebra finch, ostrich, crocodile and human
To identify potential targets of miR-2954, we used TargetScan26, which detects 6mer, 7mer-1a, 7mer-m8 and 8mer-1a target sites in the 3′ UTRs of mRNA transcripts, aligning them with the miRNA seed sequence. We obtained 3′ UTR sequences for all splice variants of genes within both the chicken (bGalGal1.mat.broiler.GRCg7b), zebra finch (bTaeGut1_v1.p), Australian saltwater crocodile (CroPor_comp1), African ostrich (ASM69896v1) and human (GRCh38.p14) genomes using BioMart (ref. 61). Subsequent identification of target sites was performed using TargetScan v.7.0 for each species. A gene was categorized as a predicted target if it contained any of these target site types within its UTRs. We then counted the total number of target sites for each predicted target gene in chicken. To calculate context+ scores, we performed a separate target prediction step specifically for chicken using TargetScan v.6.0, along with its associated Perl script (targetscan_60_context_scores.pl). We used the same chicken 3′ UTR sequences in this step as in the initial TargetScan analysis. Finally, we calculated the median context+ score for all target sites within each gene, considering only those with a context+ score of less than 0 (Supplementary Table 3).
Identification of conserved target sites in chicken and zebra finch
To identify conserved target sites, we selected the longest annotated 3′ UTR for each gene in both the chicken (G. gallus) and zebra finch (Taeniopygia guttata) genomes. These 3′ UTR sequences were then aligned using Clustal Omega. We subsequently used TargetScan v.7.0 to predict conserved target sites within the aligned sequences (Supplementary Table 3).
Differential gene expression analysis
The 3′ UTR is specific to protein-coding genes, and miRNA targets are predicted on the basis of the presence of target sites within their 3′ UTRs. Consequently, we limited the DESeq2 dataset to protein-coding genes (as identified in the GTF annotation). Differential expression analysis was conducted using DESeq2. Differentially expressed genes were identified using a threshold of less than 0.05 for Padj. according to the Benjamini–Hochberg method62. The effect of genotype on gene expression in E2 whole embryos was independently analysed in male and female embryos (model: gene expression as a function of genotype). For each tissue (head, heart and body), gene expression analysis was performed collectively across ages using a model that included both genotype and embryonic age as variables (model: gene expression ≈ genotype + embryonic age). The log fold changes and differentially expressed genes were determined for each genotype contrast (Supplementary Table 4).
Differential miRNA expression analysis
Differential expression analysis was conducted using DESeq2. Differentially expressed miRNAs were identified using a threshold of less than 0.05 for Padj. according to the Benjamini–Hochberg method62. The effect of genotype on miRNA expression in E5 head, heart and body was independently analysed in each tissue (model: gene expression as a function of genotype) (Supplementary Table 5).
Comparison of pure Hy-Line Brown females with ZW G2
Although all chickens used in the gene expression analysis were of the Hy-Line Brown breed, the G2 animals, comprising genotypes ZW, ZKOW, ZKOZ and ZKOZKO, originated from different parents compared with the ZZ genotype, which was derived from the pure Hy-Line Brown breed (the original stock). To ensure the rigour of all expression comparisons, we aimed to confirm that the G2 ZW and pure Hy-Line Brown ZW had similar gene expression profiles (ZZ embryos cannot be derived from the G2 (hemizygous/heterozygous KO) parents), thereby eliminating potential confounding factors, such as maternal effects on gene expression. Accordingly, we conducted different expression analyses between pure Hy-Line Brown ZW and G2 ZW chickens and compared the fold changes across different gene categories. This analysis confirmed that gene expression patterns are statistically indistinguishable between Hy-Line Brown and G2 and therefore do not confound our results (Extended Data Fig. 3b).
Identifying ohnologues
The list of chicken ohnologues was retrieved from the OHNOLOGS v.2 database63, available at http://ohnologs.curie.fr/ (‘relaxed’ dataset). These ohnologues were identified using gene IDs from the galGal4 assembly (Ensembl release 80), which is incompatible with the gene IDs of the chicken genome assembly used in our study (GRCG7b). To resolve this, we retrieved the unspliced DNA sequences of these ohnologue gene IDs from the GRCg6a assembly (Ensembl release 106) through BioMart. Subsequently, these sequences were aligned to the unspliced DNA sequences of protein-coding genes from the GRCG7b assembly using BLASTn (BLAST+ 2.4)64, with the settings -perc_identity 95 and -evalue 0.001. We sorted the results by bit scores to identify the best hits between the two gene sets. Cross-referencing protein names for matched gene IDs confirmed a high accuracy (88.6% exact matches) of this ID conversion method (Supplementary Table 3).
Dosage sensitivity scores
Dosage sensitivity scores for human genes, including haploinsufficiency (pHaplo) and triplosensitivity (pTriplo), were sourced from a previous study32. These scores were then assigned to chicken genes on the basis of their 1:1 orthology relationship (retrieved using BioMart) (Supplementary Table 3).
Assessment of time and tissue specificity
To evaluate the time and tissue specificity of chicken genes, we calculated time and tissue specificity indexes on the basis of the tau metric65 using a developmental time-series RNA-seq dataset34 (Supplementary Table 6). As in previous studies34, for the tissue specificity index, the tau metric was applied to the maximum expression of the gene observed during development in each organ, whereas for the time specificity index, the tau metric was applied to the expression of the gene at different time points instead of organs. In both cases, indexes range from 0 (indicating broad expression) to 1 (indicating restricted expression).
Identification of developmentally expressed genes and female-to-male expression level ratios
Gene expression ratios between the sexes were analysed using a published RNA-seq time-series dataset34,66,67. We obtained raw read (FASTQ) files for various chicken organs (blastoderm, brain, cerebellum, gonads, heart, kidney and liver) across different embryonic stages (E0, E4.5 and E6 for gonads and E10, E12, E14 and E17) and post-hatch periods (P0, P7, P35, P70 and P155). Reads were aligned to the bGalGal1.mat.broiler.GRCg7b reference genome, with read counts generated as detailed in the ‘Estimation of gene expression levels’ section. We then calculated the FPKM values for each gene using the fpkm function in DESeq2 and determined the median expression values for all embryonic and post-hatch samples (Supplementary Table 6). An FPKM threshold greater than 1, on the basis of the median for each group, was applied to filter out non-expressed and lowly expressed genes in both sexes. To identify developmentally expressed genes, we selected genes with FPKM greater than 1 in at least one tissue and time point (Supplementary Table 3).
Assessment of Z to proto-Z expression levels
For this analysis, RNA-seq data (log2-transformed reads per kilobase of transcript per million reads mapped values from ref. 34) from brain, cerebellum, heart, kidney and liver from adult male and female chicken (P155), and the corresponding stage in mice (P63) was used. Akin to previous studies5,6,9, ancestral expression levels of Z-linked genes (proto-Z genes) were estimated by calculating the median expression levels of the corresponding expressed autosomal 1∶1 orthologues in an outgroup species with non-ZW sex chromosomes (in this case, mouse). In a similar way, ancestral expression levels of autosomal genes (proto-autosomal genes) were estimated by calculating the median expression levels of corresponding 1∶1 orthologues that are autosomal in the same outgroup species with non-ZW sex chromosomes.
To obtain the current-Z to proto-Z expression ratios, we first normalized the current expression levels of Z-linked genes by the median current expression level of all 1∶1 orthologous genes that are autosomal in the outgroup species. We then normalized the ancestral expression levels of each proto-Z-linked gene (computed as described above) by the median ancestral expression level of all proto-autosomes in the outgroup species. We then computed the ratio of these two values for each gene, resulting in the current-Z to proto-Z ratios.
Finally, we compared the current-Z to proto-Z ratios for Z-linked miR-2954 targets and Z-linked miR-2954 non-targets. As Z-linked targets, we used the experimental miR-2954 targets; as non-targets, we used Z-linked genes that are neither experimental miR-2954 targets nor predicted miR-2954 targets. In both cases, we made sure that autosomal miR-2954 targets were excluded when normalizing the expression of current-Z and proto-Z genes by current-autosomal and proto-autosomal genes. Statistically significant deviations of the medians of these ratios from key reference values (for example, 0.5 (log2 ratio of −1), 1 (log2 ratio of 0) and 2 (log2 ratio of 1)) were assessed using one-sample Wilcoxon signed-rank tests. P values were corrected for multiple testing using the Bonferroni procedure68, with Padj. < 0.05 indicating significance. Statistical equivalence to these same reference values was assessed using Wilcoxon TOST (two one-sided test) equivalence tests. This approach tests whether the medians fall within a predefined equivalence margin around each reference value, meaning the expression ratios are neither significantly above nor significantly below the specified bounds. In this analysis, the equivalence bounds were set as the reference value ± 0.5. P values were corrected using the Benjamini–Hochberg procedure, with Padj. < 0.05 on both one-sided tests required for significant equivalence.
Location of genes along the Z chromosome
To visualize the location of target genes on the Z chromosome, we counted the number of protein-coding genes in windows of 0.5 Mb on the basis of gene annotations of Ensembl57 (v.111). To indicate the location of the MHM regions, we used the regions defined by Sun et al.47. We lifted these regions from Galgal5.0 to the bGalGal1.mat.broiler.GRCg7b genome assembly by extracting flanking sequences from and aligning them to the new genome with BLAT.
Sequence conservation
The sequence of the miR-2954 locus was retrieved from the National Center for Biotechnology Information (NCBI) and blasted against the reference genomes of the target species (Extended Data Fig. 9 and Supplementary Data 3) using BLASTn64.
RNA isolation, reverse transcription and RT–qPCR for miR-2954
Total RNA, including miRNA, was isolated from seven tissues (bursa of Fabricius, leg bone, brain, heart, intestine, liver and pectoral muscle) of six individual E12 chicken embryos (three male and three female; Lohmann breed) using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Approximately 50 mg of each tissue was homogenized in 500 µl of TRIzol using a TissueLyser LT (QIAGEN) at 40 Hz for 1–2 min. RNA quality was assessed by visualizing the 28S and 18S ribosomal RNA (rRNA) bands on a denaturing agarose gel and further quantified using a NanoDrop spectrophotometer.
Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) in accordance with the manufacturer’s instructions. In each 15-µl reaction, 1,000 ng of total RNA was reverse transcribed using stem-loop reverse transcription primers specific for gga-miR-2954 (Assay ID: 243071_mat; Applied Biosystems) and U6 small nuclear RNA (snRNA) (Assay ID: 001973; Applied Biosystems). The cycling conditions were as follows: 16 °C for 30 min, 42 °C for 30 min and 85 °C for 5 min, followed by holding at 4 °C.
qPCR was performed using TaqMan MicroRNA Assay for gga-miR-2954 (Assay ID: 243071_mat) and U6 snRNA (Assay ID: 001973) as the endogenous control for normalization on a QuantStudio 6 Real-Time PCR System (Applied Biosystems) following the manufacturer’s protocol. Each sample was run in triplicate. Each 10-µl reaction mixture contained 0.66 µl of complementary DNA (cDNA), 0.5 µl of TaqMan MicroRNA Assay and 5 µl of TaqMan Fast Advanced Master Mix (catalogue no. 4444557). The cycling conditions were as follows: 95 °C for 20 s, followed by 40 cycles of 95 °C for 1 s and 60 °C for 20 s.
The cycle threshold values were normalized using the 2ΔCT method, where ΔCT is the difference between the target gene and the endogenous control (U6 snRNA) (Supplementary Table 8).
miR-2954 knockdown and RT–qPCR
miR-2954 knockdown was achieved by injecting mirVana miRNA inhibitor specific to miR-2954 (Thermo Fisher Scientific; catalogue no. 4464088) or mirVana miRNA Inhibitor, Negative Control #1 (Thermo Fisher Scientific; catalogue no. 4464076) into chick embryos at two different embryonic stages. Lyophilized miRNA inhibitors (250 nmol; high-performance liquid chromatography; in vivo ready) were resuspended in nuclease-free water to prepare a stock solution with a final concentration of 2.5 mg ml−1. The miRNA inhibitor solutions were then complexed with Invivofectamine 3.0 Reagent (Thermo Fisher Scientific; catalogue no. IVF3001). The Invivofectamine 3.0–miRNA duplex mixtures were incubated for 30 min at 50 °C and subsequently diluted with PBS (pH 7.4) according to the manufacturer’s instructions.
A total of 240 fertilized eggs were obtained from Lohmann Sverige AB and placed in an incubator at 37.5 °C with 50% humidity. At E2.5, a small window was created in the eggshell above the embryo using an engraving machine. Using a fine glass needle, 2 µl of the Invivofectamine 3.0–miRNA duplex mixture (containing a final concentration of 0.63 mg ml−1 of the inhibitor) was injected into the dorsal aorta. Following successful injections in 170 knockdown and 28 negative control embryos, the eggs were sealed with tape and returned to the incubator at 37.5 °C with 50% humidity. A second injection was performed at E4 in surviving embryos using the same procedure but with 3 µl of the Invivofectamine 3.0–miRNA duplex mixture. Embryo viability was evaluated at E12 by observing blood flow after removal of the chorioallantoic membrane. A subset of embryos was frozen at E5 for subsequent gene expression analysis.
For gene expression analysis, 20 embryos (12 knockdown and eight control) were injected as described above and snap-frozen at E5, 1 day after the second injection, for subsequent RNA extraction and quantification (Supplementary Table 2 and Extended Data Fig. 5).
To determine the impact of miR-2954 knockdown on the expression of target and non-target genes, including XPA, we performed molecular sexing, dissected the heart tissue and isolated total RNA. Three controls and five knockdown embryos were used for gene expression analysis. RNA extraction was performed using TRIzol reagent, following the protocol outlined in the previous section.
Using the NCBI Primer-BLAST tool, we designed forward and reverse primers for eight target genes, eight non-target genes, the XPA gene and the reference gene GAPDH (Supplementary Table 1). Then, 1,000 ng of RNA from each sample was reverse transcribed into cDNA using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific; catalogue no. K1612) and oligo(dT) primers, according to the manufacturer’s instructions. QPCR was carried out on a QuantStudio 6 Real-Time PCR System (Applied Biosystems) using SYBR Green Universal Master Mix (catalogue no. 4309155). The thermal cycling profile consisted of an initial denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s (denaturation) and 55 °C for 60 s (annealing and extension). Each PCR was run in triplicate. A final melting curve analysis was performed to confirm the specificity of the PCR products. Data were analysed using the delta–delta cycle threshold method. The cycle threshold values were normalized to GAPDH, and log2 fold changes between miR-2954-KD and control were generated (Supplementary Table 8).
Generation of Ribo-seq data
To compare transcriptome versus translatome patterns, we used a recently developed Ribo-seq procedure69, on the basis of previously established methods27,70, optimized for generating high-quality data from low-input frozen tissue samples, including small embryonic specimens. Using this method, we generated Ribo-seq and matched RNA-seq data for a total of eight adult chicken and mouse brain (forebrain/cerebrum) samples, as well as chicken embryonic head samples (Supplementary Table 9). These data were further complemented by our previously published Ribo-seq dataset6, which cover three additional adult chicken and mouse brain samples. Detailed protocols for the new Ribo-seq and matched RNA-seq experiments are provided below, followed by a description of the methods used to analyse these data.
Ribo-seq footprint generation
Frozen tissues were lysed in 150 µl of ice-cold lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1% (v/v) Triton X-100, 1 mM dithiothreitol, 0.4 U ml−1 RiboLock and 100 µg ml−1 of cycloheximide) using a micropestle. Lysates were clarified by centrifugation at 20,000g for 7 min at 4 °C. For nuclease digestion, 450 U RNase I (Ambion) and 3.75 U TURBO DNase I (Thermo Fisher Scientific) were added, and samples were incubated at 25 °C for 45 min with gentle agitation. Digestion was stopped by the addition of 0.5-µl SUPERase·In RNase Inhibitor (Ambion).
To purify ribosome-protected fragments, lysates were overlaid on 700 µl of 30% sucrose cushion in 13 × 51 mm centrifuge tubes (Beckman Coulter). Samples were centrifuged at 100,000 rpm for 1 h at 4 °C using an S100-AT6 rotor (Ultracentrifuge Sorvall Discovery M120 SE). The supernatant was discarded, and the pellet was resuspended in 700 µl of 10 mM Tris (pH 7.0). To extract RNA, 40 µl of 20% sodium dodecyl sulfate and 750 µl of 65 °C acid phenol:chloroform were added, followed by incubation at 65 °C for 10 min with agitation. After centrifugation at a maximum speed for 4 min, the aqueous phase was transferred to a fresh tube containing 700 µl of acid phenol:chloroform, incubated at room temperature with intermittent vortexing and centrifuged for 4 min. Next, 600-µl chloroform was added, vortexed and centrifuged for 4 min. RNA was precipitated overnight at −70 °C in the presence of 600-µl isopropanol, 66.7 µl of 3 M sodium acetate (pH 5.5) and 2-µl GlycoBlue (Thermo Fisher Scientific). RNA was pelleted by centrifugation for 40 min at maximum speed, washed with 80% ethanol and resuspended in 12.5 µl of 10 mM Tris (pH 7.0).
The extracted RNA was separated on a 15% denaturing urea polyacrylamide gel (Thermo Fisher Scientific) and stained with SYBR Gold (Thermo Fisher Scientific). Fragments of 27–33 nt were excised and disrupted using gel breaker tubes. RNA was extracted in 0.5 ml of 10 mM Tris (pH 7.0) for 10 min at 70 °C with agitation. Gel debris was removed by centrifugation in Spin-X filter tubes (Corning) for 2 min at maximum speed. RNA was precipitated overnight at −70 °C in the presence of 1 volume isopropanol, 0.1 volume 3 M sodium acetate (pH 5.5) and 2-µl GlycoBlue (Thermo Fisher Scientific). RNA was pelleted by centrifugation for 40 min at maximum speed and washed with 80% ethanol.
Ribo-seq library preparation and sequencing
Ribo-seq library preparation was performed as described in ref. 69 with several modifications. In brief, ribosome footprints were dephosphorylated and ligated to a pre-adenylated 3′ linker (L1), followed by enzymatic removal of unligated linkers. Footprint–linker complexes were captured on streptavidin beads, phosphorylated and ligated to a 5′ linker (L2). Reverse transcription was performed on bead-bound templates, and the resulting cDNA libraries were amplified by PCR. To improve depletion of unligated L1, we modified the digestion step by incubating samples sequentially at 30 °C for 60 min and 37 °C for 60 min with deadenylase and RecJf. Libraries were PCR-amplified using eight cycles of amplification. A modified version of the previously published Cas9-mediated Ribocutter tool71 was used to deplete rRNA from the Ribo-seq libraries. The sgRNAs were designed to target the most abundant contaminants of previously sequenced libraries derived from chicken or mouse telencephalon. To enhance the efficiency of rRNA removal, a lower library concentration (6 nM) was used as input for Cas9-mediated depletion and extended the Cas9 treatment to 4.5 h. An additional 6% polyacrylamide gel electrophoresis Tris–borate–EDTA gel step was introduced to remove preferentially amplified adaptor dimers following a seven-cycle PCR reamplification. All further steps were performed according to the original protocol. The libraries were resuspended and quality controlled using Qubit (Thermo Fisher Scientific) and Fragment Analyzer (Agilent Technologies) platforms. Sequencing was performed on both the Illumina NextSeq 2000 and Illumina NextSeq 550 systems, using NextSeq 2000 P4 Reagents (50 cycles) and NextSeq 550 High Output Reagents (75 cycles). The samples were multiplexed into one set of six and another set of one. All further steps were performed in accordance with the original protocol.
RNA library preparation and sequencing
To generate matched RNA-seq libraries prepared from the same lysates, total RNA was extracted from dissected tissues using the RNeasy Micro Kit (QIAGEN), following the manufacturer’s instructions. The RNA quality was assessed using the Fragment Analyzer system (Agilent), and RNA quality numbers ranged from 7.7 to 10, indicating minimal degradation. The RNA-seq libraries were prepared using the SMART-Seq Total RNA High Input kit with (Mammalian) RiboGone (Takara Bio). The concentration and quality of the libraries were determined using Qubit (Thermo Fisher Scientific) and Bioanalyzer (Agilent Technologies) platforms. Illumina sequencing was performed on an Illumina NextSeq 2000 system using NextSeq 2000 P2 Reagents (100 cycles), with samples multiplexed in one set of six.
Read mapping and processing
Raw sequencing reads with Illumina 3′ adaptor and low-quality bases (Phred score below 20) were trimmed using cutadapt v.4.6 (parameters: –adapter=AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC –minimum-length=6 -q 20). For Ribo-seq libraries, unique molecular identifiers (UMIs) were extracted using UMI-tools v1.1.4 (ref. 72) (parameters: –bc-pattern = ^(?P<umi_1 > .{5}). + (?P<umi_2 > .{5})$ –extract-method=regex OR –bc-pattern = ^(?P<umi_1 > .{10}). + (?P<umi_2 > .{10})$ –extract-method=regex), and leading nucleotides were removed with cutadapt v.4.6 (parameters: -u 6). RNA-seq libraries did not contain UMIs, and leading nucleotides were removed with cutadapt v.4.6 (parameters: -u 3). Trimmed reads were consecutively mapped to the index libraries of species-specific (chicken or mouse) contaminating RNAs obtained from RNAcentral73 (rRNAs, mitochondrial RNAs and transfer RNAs) using Bowtie 2 v.2.5.1 (ref. 74) (parameters: –phred33 -L 20 -N 1 -t –no-unal). Aligned reads were discarded, and only those within the defined length ranges (26–34 nt for Ribo-seq and 20–50 nt for RNA-seq) were kept for downstream analysis. As expected27, Ribo-seq read lengths peaked at 28–30 nt and predominately mapped to coding DNA sequences (CDSs) (Extended Data Fig. 4a,b). To mitigate bias in the mapping of RNA-seq reads in exon–exon junctions owing to length discrepancies between both methods, RNA-seq reads were cut to 29 nt. Reads were then aligned to the reference genomes (bGalGal1.mat.broiler.GRCg7b; GCA_016699485.1 OR GRCm39; GCA_000001635.9, Ensembl release 113; ref. 75) using STAR aligner v.2.7.11a (ref. 58) (parameters: –alignEndsType EndToEnd –outSAMattributes All –outSAMtype BAM SortedByCoordinate –outMultimapperOrder Random). As previously described27, peptidyl-site offsets were estimated per read length, and Ribo-seq reads were calibrated accordingly.
Triplet periodicity
To assess whether our Ribo-seq libraries showed patterns of true translation, we analysed the triplet periodicity using raw reads mapped to the complete CDS regions of protein-coding genes. To ensure robust analysis, we focused on protein-coding genes annotated as canonical in Ensembl (release 113). The number of reads mapped to the three reading frames was normalized by the total number of reads within the CDS. As shown in Extended Data Fig. 4c,d, in contrast to RNA-seq reads, our Ribo-seq data predominantly mapped to the first nucleotide of the codon showing continuous and significant triplet periodicity across the CDS.
Estimation of gene expression levels
Transcript abundances were estimated in FPKM. Only uniquely mapped RNA-seq reads and de-duplicated uniquely mapped Ribo-seq reads within the CDS regions were considered. On the basis of our triplet periodicity analysis, we further restricted the analysis to read lengths that exhibited significant triplet periodicity. Moreover, only the CDS region from the +4th to the −3rd codon was used to avoid inflated counts owing to random translation initiation and ribosome enrichment at the stop codon70. For each gene, the longest isoform was used as a representative. Gene count matrices were then loaded into R v.4.4.0, and gene expression levels were estimated using the rpkm function of edgeR v.4.2.0, which accounts for both CDS length and library depth. The FPKM values were log2-transformed.
Assessment of reproducibility
To assess the reproducibility of our Ribo-seq data, we calculated the Spearman’s correlation coefficient (ρ) between the read counts of canonical protein-coding genes in two biological chicken female brain replicates. The high correlation (ρ = 0.98) demonstrates strong biological reproducibility (Extended Data Fig. 4e).
Comparison of ZKOZKO and ZZ genotypes
Processed FPKM values were used to calculate log2[FC] in gene expression between ZKOZKO and ZZ genotypes for both layers (transcriptome and translatome) in head tissue. Genes with FPKM values greater than 1 in both genotypes were kept to exclude non-expressed or lowly expressed genes. To enable direct comparisons between layers, the FPKM values were normalized using the median expression of autosomal non-target transcriptome or translatome genes, respectively.
Assessment of Z to proto-Z translation levels
To estimate the ancestral translatome levels of Z-linked genes, we combined our newly generated Ribo-seq data with our previously published dataset6. The analysis followed the same approach as the RNA-seq analysis described earlier (Assessment of Z to proto-Z expression levels).
Female-to-male expression-level ratios
The processed FPKM values were used to calculate female-to-male ratios for two tissues (fetal head and adult brain) for both layers (transcriptome and translatome). Genes with FPKM values greater than 1 were kept to filter out non-expressed and lowly expressed genes. To allow for comparisons between layers, the FPKM values were normalized using either the median of autosomal transcriptome or translatome expression. The ratios were then compared to the key reference values, as described earlier (Assessment of Z to proto-Z expression levels).
Translation efficiency estimation
The log2-transformed FPKM values at the translatome (ribosome-protected fragment) and transcriptome (RNA) were used to calculate translation efficiency across samples as:
$${\rm{TE}}={\log }_{2}({{\rm{RPF}}}_{{\rm{FPKM}}})-{\log }_{2}({{\rm{RNA}}}_{{\rm{FPKM}}})$$
where RPF is the ribosome-protected fragment, and TE is the translation efficiency. Further, to highlight the differences between male and female translation efficiencies, the female-to-male-translation-efficiency ratios were calculated as:
$${{\rm{TE}}}_{{\rm{F}}-{\rm{to}}-{\rm{M}}}={\log }_{2}({{\rm{TE}}}_{{\rm{female}}})-{\log }_{2}({{\rm{TE}}}_{{\rm{male}}})$$
Finally, the ratios were normalized using the median of autosomal female-to-male-translation-efficiency ratios.
Long-read genome sequencing of miR-2954 KOs and controls
For long-read sequencing, we selected five miR-2954 KO individuals and four non-edited controls. Genomic DNA was isolated through DNeasy Blood & Tissue Kit (QIAGEN). Library preparation was performed using the Rapid Barcoding Kit (SQK-RBK114-24) or the Native Barcoding Kit (SQK-NBD114-24) (Oxford Nanopore Technologies). Sequencing was conducted on PromethION R10.4.1 flow cells with adaptive sampling76 to specifically enrich for Z-chromosomal reads.
For basecalling, we used the high-accuracy model ([email protected]) implemented in dorado-0.9.0. Reads were then aligned to the GRCg7b chicken genome assembly using minimap2 (ref. 77) (v.2.27-r1193) with the long-read high-quality preset (-x lr:hq). The per-sample coverage depth across chromosome Z was calculated using SAMtools 1.20 (ref. 78) depth command.
We next searched for structural variants using Sniffles2 (v.2.2)79, configuring the tool to call small indels and putative structural variants on chromosome Z with the parameters –minsvlen 5 and –minsupport 0. This analysis identified six candidate variants shared among the five KO samples but absent in controls (Supplementary Table 10). Manual inspection of these variants in Integrative Genomics Viewer revealed that only one site showed a consistent difference between KO and control samples: the intended Cas9 target, which produced a 32-bp deletion starting at chr. Z: 71,305,198. Notably, this deletion was also the only variant displaying zero coverage exclusively in KO individuals, confirming the successful miR-2954 KO.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.