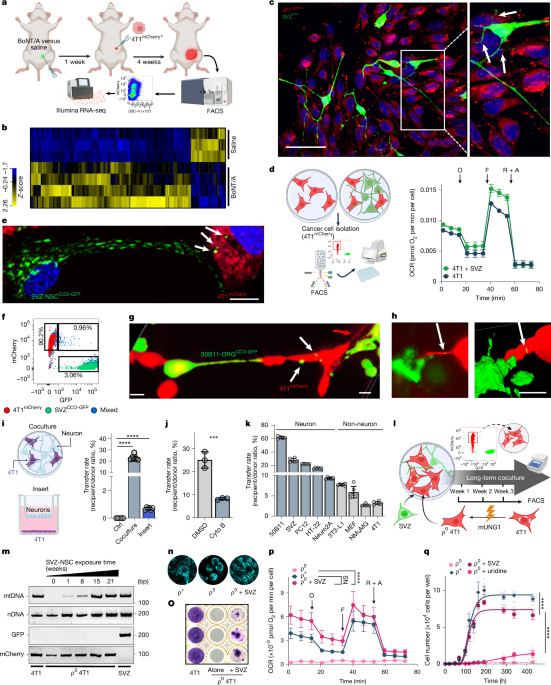
Cell culture
The 4T1 (CRL-2539), Neuro2A (CCL-131) and NMuMG (CRL-1636) cell lines were purchased from ATCC. HT-22 (ESA111) and 3T3-L1 (EF3001) were purchased from Kerafast. The 50B11 rat DRG neuronal cells were obtained from the GRCF Biorepository & Cell Center (Johns Hopkins University). MEFs were provided by S. Lloyd. Primary SVZ-NSCs were isolated from fresh BALB/c mouse brains as described previously60 (Extended Data Fig. 2a–d). The cell lines were routinely checked for mycoplasma contamination by using the Mycostrip Test Kit (Invivogen), and through Hoechst-33342 staining. The cell lines used on this study has been authenticated by genomic profiling. Cancer cells were cultured using Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 5% fetal bovine serum (FBS; Fisher Scientific), 5% bovine calf serum (Fisher Scientific), antimicrobial solution (penicillin, streptomycin and amphotericin B; 1% antibiotic–antimycotic, Life Technologies) and antimycoplasma solution (1.25 mg l−1 plasmocin prophylactic, InvivoGen). SVZ-NSCs were cultured in specialized medium (Neurobasal, Life Technologies) supplemented with serum-free supplement (2% B-27, Life Technologies), 1 mg l−1 heparin (STEMCELL Technologies), l-alanyl-l-glutamine dipeptide supplement (1% GlutaMAX, Life Technologies), antimicrobial solution (1% antibiotic–antimycotic, Life Technologies), 10 ng ml−1 recombinant human epidermal growth factor (Bio-Techne) and 10 ng ml−1 of recombinant human fibroblast growth factor basic (Bio-Techne). 50B11 cells were cultured in specialized medium (Neurobasal, Life Technologies) supplemented with both 10% serum and the serum-free supplement (2% B-27, Life Technologies), l-alanyl-l-glutamine dipeptide supplement (0.27% GlutaMAX, Life Technologies), antimicrobial solution (1% antibiotic–antimycotic, Life Technologies) and 0.22% glucose. SVZ-NSC adherence in vitro was aided by adding 0.25 µl ml−1 ECMatrix-511 Silk E8 Laminin Substrate (MilliporeSigma). For serum-free experiments, FBS and bovine calf serum were substituted with a serum-free medium (10% Knockout Serum Replacement (KSR), Life Technologies). Dialysed FBS (Thermo Fisher Scientific) was substituted for FBS in uridine depletion assays. ρ0 cells were cultured in medium supplemented with 50 µg ml−1 uridine (Sigma-Aldrich) and 1 mM sodium pyruvate (Fisher Scientific). Antibiotic selection pressure was applied with combinations of 10–20 µg ml−1 puromycin (Fisher Scientific), 100–200 µg ml−1 phleomycin D1 (Zeocin, InvivoGen) and 10–20 µg ml−1 Blasticidin (InvivoGen) to maintain transgene expression. Cells were cultured in an incubator at 37 °C and 5% carbon dioxide and were tested routinely for mycoplasma contamination.
In short-term nerve–cancer coculture experiments (≤5 days), 4T1 cells were initially seeded in DMEM and allowed to adhere. After 1 day, the medium was changed to KSR medium, and SVZ-NSCs were added the following day. For long-term coculture experiments, 4T1 cells were maintained in DMEM, with SVZ-NSCs added weekly and 4T1 cells passaged twice a week. SVZ-NSCs were introduced into the 4T1 cell cultures at 2–4% of the total cell population, matching the percentage observed in primary cancers formed from 4T1 cell xenografts61. For the 3D coculture and reconstruction imaging experiment, a total of 1.2 × 105 50B11CCO-GFP cells and 1.2 × 105 4T1mCherry cells (both in 10 µl PBS) were mixed with 60 µl of Growth Factor Reduced Matrigel (Corning, catalogue number 356231) and seeded onto the glass area of uncoated glass-bottomed 35-mm dishes (MatTek). The dishes were incubated at 37 °C in a humidified atmosphere of 5% CO2 in air for 1 h. Following incubation, 2 ml of Neurobasal complete medium containing 75 µM forskolin (MilliporeSigma) was added. Confocal images were captured 2 days later using a Nikon A1 confocal laser microscope.
Virus production and cell line generation
Lenti-X 293T cells (Takara Bio USA) were used for lentivirus generation, and Phoenix-AMPHO cells (ATCC) were used for retrovirus production. Cells were plated in complete DMEM 1 day before transfection using a transfection reagent (FuGENE, 3 µl µg−1 of plasmid; Promega).
For lentivirus production, Lenti-X 293T cells were transfected by lipofection with the following components: 1,000 ng of transfer plasmid, 750 ng of packaging plasmid (psPAX2; Addgene) and 250 ng of vesicular stomatitis virus G envelope-expressing plasmid (pMD2.G; Addgene). Both psPAX2 and pMD2.G were gifts from D. Trono, École Polytechnique Fédérale de Lausanne (Addgene plasmid numbers 12260 and 12259). For in vivo work, the lentivirus was further concentrated in a concentrator solution (40% PEG-8000; 1.2 M NaCl in PBS) added at a 1:3 ratio of solution/supernatant, gently shaken for 2 days at 4 °C, collected through 1,600g centrifugation and resuspended in a 1:100 volume of PBS and stored at −80 °C.
For retrovirus production, Phoenix-AMPHO cells were transfected with 1,000 ng of plasmid DNA. The medium was replaced with KSR-containing medium 1 day after transfection, and the viral supernatant was collected over a period of 2 days. For viral transduction, target cells were plated at 60% confluence and transduced in the presence of 10 µg ml−1 Polybrene (MilliporeSigma) in KSR medium. SVZ-NSCs were cultured in Neurobasal medium without Polybrene to maintain cell viability. The pLenti-CMV-GFP-puro plasmid was provided by E. Campeau (Addgene plasmid number 17448). The pLVX-EF1a-CCO-IRES-puromycin plasmid was provided by D. Andrews (Addgene plasmid number 134861). The pFUGW mCherry-KASH plasmid was a gift from H. MacGillavry (Addgene plasmid number 131505). The CMV-loxP-DsRed-loxP-eGFP plasmid was provided by D. Gilkes (Addgene plasmid number 141148). The pLenti-CMV-puro-2A-TEVp plasmid was a gift from M. Tripodi (Addgene plasmid number 99610).
ρ0 4T1 cell lines were developed as described previously28. The complete and permanent loss of mtDNA in 4T1 cells was achieved by transient co-overexpression of a mUNG1 and UL12.5M185 herpesvirus protein. 4T1mCherry cells were cultured on 6-cm plates. At 1 day after seeding, cells were lipofected using 15 µl of FuGENE transfection reagent and 2,500 ng each of pMA4008 and pMA3790 plasmids provided by M. Alexeyev62. Lipofection was performed on medium with uridine and sodium pyruvate and without antibiotics, and medium was changed after 1 day to fresh medium with uridine, sodium pyruvate and antibiotics. Single-cell sorting based on strong positive eGFP expression (top 3%) was performed after 3 days into 96-well plates supplemented with uridine, sodium pyruvate and 20% FBS. The medium was changed weekly and twice weekly when colonies were observed.
The MitoTRACER donor cells were prepared using retroviral or lentiviral transduction, followed by selection with 20 µg ml−1 Blasticidin. To further purify cells with the highest MitoTRACER expression, we incorporated a GFP-11 strand into the MitoTRACER construct, allowing for additional selection by FACS. If necessary, the MitoTRACER cells were further purified to ensure consistent and robust expression by transiently transfecting them with 5,000 ng of GFP1-10 plasmid and sorting based on GFP signal 3 days later. The plasmid pQCXIP-GFP1-10 was provided by Y. Hata (Addgene plasmid number 68715). Loss of GFP signal following transient selection was confirmed by flow cytometry before utilizing the cells in MitoTRACER coculture experiments.
DNA extraction and PCR
DNA was extracted from cells using a kit (GeneJET Genomic DNA Purification Kit, Thermo Fisher Scientific). Reaction mixes were used for end-point PCR (DreamTaq Hot Start Green PCR Master Mix, Thermo Fisher Scientific) and quantitative PCR (PowerTrack SYBR Green Master Mix, Thermo Fisher Scientific). The quantitative PCR was performed with a thermocycler (QuantStudio 7 Pro, Thermo Fisher Scientific). Sample concentration was adjusted to 10 ng µl−1, and PCR was performed with 20 ng of DNA template. The end-point PCR products were separated on acrylamide/bis-acrylamide (29:1) 10% gels (Thermo Fisher Scientific) and stained with ethidium bromide (Thermo Fisher Scientific). Mouse mtDNA amplification was performed as described previously63 using the mMitoF1 5′-CTAGAAACCCCGAAACCAAA-3′ and mMitoR1 5′-CCAGCTATCACCAAGCTCGT-3′ primers. The mtDNA copy numbers were normalized to the nuclear copy numbers using the B2MF1 5′-ATGGGAAGCCGAACATACTG-3′ and B2M-R1 5′-CAGTCTCAGTGGGGGTGAAT-3′ primers. Uncropped gels are provided in Supplementary Fig. 1.
Western blot
Western blot was performed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Whole-cell lysates were prepared from 2–5 × 106 cells in 300 µl of lysis buffer (20 mM Tris, pH 7.4, 1% Triton X-100, 10% glycerol, 137 mM sodium chloride, 2 mM ethylene diamine tetraacetic acid, 1 mM sodium orthovanadate) and protease inhibitors (Thermo Fisher Scientific). Subcellular fractionation was performed using the Mitochondria Isolation Kit for Cultured Cells (Life Technologies) according to the manufacturer’s instructions. With mitochondrial-bound proteins, sonication (30 pulses, each 20 s) was performed instead of centrifugation of excess material to ensure that mitochondrial proteins remained in suspension. Lysates were clarified by centrifugation at 4 °C for 30 min at 17,000g. Typically, whole-cell lysates (5–20 µg) were separated on 10% or 12% acrylamide minigels and transferred to a membrane (Immuno-Blot, Bio-Rad).
Samples were separated with electrophoresis (Any kD gels and Mini-PROTEAN apparatus; Bio-Rad). The membrane was blocked for 30 min in wash buffer (0.1% Tween 20 in PBS) containing 5% nonfat dry milk and incubated overnight with primary antibody (Cre recombinase (1:1,000, number 15036, lot 2), mCherry (1:1,000, number 43590, lot 2) or α-tubulin (1:1,000 number 2144, lot 5), Cell Signaling Technology; or GFP (1:200, number sc-9996, lot E2521) or HA tag (1:1,000, number sc-7392 lot I0992), Santa Cruz Biotechnology) that was diluted in the same buffer. After extensive washing, the blot was incubated with secondary antibody (1:1,000, Thermo Fisher PI31430 or PI31460) for 30 min in blocking buffer, washed and processed with a western blot detection system (ChemiDoc MP Imaging System, Bio-Rad). Uncropped blots are provided in Supplementary Fig. 1.
Imaging and flow cytometry analysis
Cell imaging was performed with a fluorescence (BZ-X810, Keyence) or inverted confocal microscope (A1R, Nikon). Cell sorting was performed using FACS (FACSAria II (BD Biosciences) with analysis of GFP (laser excitation, 488 nm; detection, 525/30 band-pass filter width (BP)) and DsRed-Express2 and mCherry Red fluorophores (561 nm; 610/20 BP). The gating strategy is provided in Supplementary Fig. 2.
Dye staining (MitoTracker, Invitrogen) was performed according to the protocol from the manufacturer. Sorting was performed with 1 cell per well in 96-well plates (single-cell sorting) or 5,000 cells per well in 6-well plates containing 2 ml of DMEM with 10% FBS. Immunostaining and cytometry were performed as follows: cells were collected by trypsinization, fixed in 4% paraformaldehyde for 15 min at room temperature, and permeabilized with 0.1% Triton X-100 diluted in 0.5% bovine serum albumin. Fixed cells were incubated for 1 h at room temperature with primary antibodies, washed in PBS and incubated for 30 min at room temperature with an Alexa Fluor 647 secondary antibody (1:10,000, Thermo Fisher A21235, lot 2836809; A21244, lot 2674387). For animal xenograft samples, deparaffinized slides were microwave-boiled (heat-induced epitope retrieval) with 10 mM sodium citrate, blocked in 1% bovine serum albumin, incubated in primary antibodies overnight, washed, incubated in secondary antibodies, Alexa 647 and Alexa 568, for 40–60 min (1:250, Thermo Fisher A78952; lot 3034155, A11004, lot 2198584) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The following primary antibodies were used: O4 (1:10,000, R&D Systems MAB1326, lot HWW143051), ALDH1L1 (1:10,000, OriGene TA501868S, lot A01), Map2 (1:1,000 Proteintech 17490-I-AP, lot 00126726), GFP (1:200, Invitrogen A10262, lot 2738237) and β3 Tubb3 (1:400, Santa Cruz sc80005, lot A1821).
Whole-cell current clamp recordings
Whole-cell current clamp recordings were performed on neurons differentiated from cocultures of SVZ-neural progenitor cell mixed with 4T1 cancer cells using an Axopatch 200B amplifier and Digidata 1322A, with data acquisition through pClamp 8 software (Molecular Devices). A Zeiss Axiovert microscope with epifluorescence was used to identify and record neural cells expressing eGFP in the nerve–cancer coculture. Cells were clamped at −80 to −90 mV, and depolarizing currents were injected using 10 current ramp increments from 20 to 200 pm (800 ms duration), interleaved by 5-s intervals. Membrane voltages were sampled at 5 kHz and filtered at 2 kHz. The action potential threshold was determined from the dV/dt derivative calculated using IgorPro 6 (WaveMetrics)64,65.
The external bath solution consisted of 120 mM NaCl, 6 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES and 20 mM glucose (pH 7.4). Patch pipettes were filled with an internal solution containing 125 mM K+ gluconate, 4 mM KCl, 4 mM NaCl, 1 mM MgCl2, 10 mM HEPES, 4 mM MgATP, 0.3 mM Na2GTP and 10 mM phosphocreatine (pH 7.26). Osmolarity of all solutions was verified with an Osmette III osmometer (Precision Systems). Series resistance and membrane capacitance were not compensated but were measured at the beginning and end of each recording to ensure quality. Electrophysiology figures were prepared using IgorPro and GraphPad Prism 9.
Uridine auxotrophy assay
The ρ0 4T1mCherry+ cells were cocultured with SVZ-NSCsCCO-GFP and sorted on the basis of mCherry and eGFP expression using flow cytometry. The sorted cells were seeded on DMEM with dialysed FBS, with or without uridine and sodium pyruvate, and stained with 2% crystal violet in 20% methanol. The rescued cells were cultured in the presence of antibiotic (Zeocin), associated with the cancer cell expression of the mCherry fluorophore. 4T1 cells were checked for the presence of mCherry and the absence of eGFP expression to confirm the 4T1 nature of the collected cells and exclude contamination with SVZ-NSCs during FACS.
Seahorse metabolic assay
Cells were plated at 4 × 104 cells per well (Seahorse XFe96 plate, Agilent) on the day before assay, and the assay tools and calibrant solution (Seahorse FluxPak, Agilent) were acclimated in an incubator without carbon dioxide on the night before assay according to the protocol from the manufacturer. An assay kit was used (Seahorse XF Mito Stress Test Kit, Agilent) with oligomycin (1.5 µM), FCCP (1.0 µM), and rotenone and antimycin A (0.5 µM). On the assay day, the medium was changed (Seahorse XF DMEM, Agilent) and supplemented with 10 mM glucose (Agilent), 1 mM pyruvate (Agilent) and 2 mM glutamine (Agilent). Uridine and sodium pyruvate were added for assays with ρ0 cells. Assays were performed according to the protocol from the manufacturer, and wells were normalized by direct cell counting to determine the absolute number of cells per well. Cells were stained with Hoechst dye and counted directly with an automatic microscope reader (Celigo S, Nexcelom Bioscience). The absence of cross-contamination following the sorting of coculture was confirmed by the analysis of the presence of eGFP+ cells in the mCherry-FACS-sorted cancer cell samples.
Mitochondria transfer assays
In cases in which inserts were used to physically block contact between cell types, 100,000 4T1mCherry cells were seeded in the bottom well of a 6-well Transwell system (Corning, 3412) or 6-well plate, and the following day, the medium was changed to 10% KSR-containing DMEM, and 200,000 SVZ-NSCsCCO-GFP were added either to the insert or directly to the well. For the dose–response and time–response assays, 4T1mCherry cells were seeded at 100,000 cells per well and changed to KSR-containing medium the next day. For the dose–response assay, between 25,000 and 400,000 SVZ-NSCsMitoTRACER were then added to the culture. For the time–response assay, 200,000 SVZ-NSCsMitoTRACER were added daily until the time of FACS analysis. FACS analysis was carried out on the third day after adding SVZ-NSCsMitoTRACER.
Donor cell cloning
The MitoTRACER construct was cloned into the pMXs-IRES-Blasticidin retroviral vector RTV-016. We used the HA-MITO plasmid pMXs-3XHA-EGFP-OMP25 as a template. pMXs-3XHA-EGFP-OMP25 was a gift from D. Sabatini (Addgene plasmid number 83356). The OMP25 mitochondrial tagging element and attached 3′ linker (Pro-Arg-His) were conserved. The HA-MITO plasmid was digested to insert our construct design consisting of the assembly of GFP-11–HA tag–TEVp cleavage recognition site–linker–iCre recombinase–SV40-NLS. The GFP-11 fragment was added between the OMP25 and TEVp cleavage site for convenience to generate cells having the desired amount of reporter using GFP1-10 fluorescence complementation and FACS. The construct also included a Blasticidin S deaminase gene separated by an internal ribosome entry site (IRES), enabling the selection of cells that were transduced successfully by the retroviruses. All constructs were validated by Nanopore sequencing. For in situ labelling, the MitoTRACER construct was transferred into a lentiviral construct with the Syn1 promoter (Addgene plasmid number 71427). The CRE-NLS control was generated through site-directed mutagenesis (Q5 Site-Directed Mutagenesis E0554S, New England Biolabs) by removing the OMP25 section of MitoTRACER. Similar constructs were generated by replacing the MitoTRACER with the eGFP fluorophore instead to generate the lentiviral Syn1-GFP-OMP25 and Syn1-GFP-NLS constructs.
Recipient cell cloning
The recipient cell construct for the MitoTRACER method was a DsRED and eGFP loxP switch with TEVp expression under the control of an SV40 promoter and tied to a BleoR gene by a 2A self-cleaving peptide. This construct was derived from the CMV-loxP-DsRed-loxP-eGFP plasmid (Addgene number 141148) that was digested with DraIII and SwaI restriction enzymes to insert the F2A-TEVp at the Ct end of BleoR with a DNA assembly kit (NEBuilder HiFi DNA Assembly, New England Biolabs).
Laser-capture microdissection, RNA extraction and cDNA microarray analysis
Laser-capture microdissection, RNA extraction and cDNA microarray analysis were conducted as previously detailed2,66. Cancer cells from BoNT/A and saline groups were procured through laser-capture microdissection from 8-μm frozen sections cut using a cryostat from the cancerous areas of fresh, unfixed xenograft specimens. The sections were mounted on non-charged glass slides and stored at −80 °C until analysis. Following staining and dehydration as per the manufacturer’s protocol, a laser-capture microdissection was performed before cDNA preparation, and microarray analyses as previously described2. After performing quantile normalization, we carried out principal component analysis as it was noted that samples from the BoNT/A and saline groups have clear distinct expression patterns. A total of 116 genes (135 probes) were identified as differently regulated by an empirical Bayes test with a false discovery rate cutoff <0.30 and a twofold change as an exploratory approach. To identify the biological processes and pathways associated with the differentially expressed genes, we performed a GATHER analysis67 and used gene set enrichment analysis68 to score association of BoNT/A treatment with Kyoto Encyclopedia of Genes and Genomes (KEGG)69 pathways.
RNA-seq of mouse mammary fat pad denervation model
Reads were trimmed for quality control to remove low-quality bases and adaptor sequences. Samples failing quality assurance and quality control thresholds (based on read quality, length or count) were excluded. Quality-trimmed reads were aligned to the Mus musculus mm39 genome using Bowtie2, in paired-end mode with a 500-bp fragment length. SAM and BAM files were processed with SAMtools and Sambamba, and alignment quality was assessed using SAMtools idxstats, retaining only reads aligning to mm39. Read counts were normalized with DESeq2’s median-of-ratios method, and genes with expression below 1.0 (geometric mean) were filtered out. Gene set ANOVA was performed on gene sets after excluding low expression levels (geometric mean <1.0) on sets containing between 2 and 50 genes. ANOVA was applied using a log-normal model with the criterion Akaike information criterion (AIC), followed by false discovery rate (FDR) correction for multiple comparisons. KEGG pathways were analysed to identify differentially regulated gene sets.
Nanopore mtDNA sequencing and classification of mtDNA heteroplasmy
To analyse mtDNA transfer in situ, cancer cells were FACS-sorted to isolate mCherry+ 4T1 cancer cells from tumour tissues. DNA was extracted from collected cells using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific). The mtDNA region containing the mutation site was amplified using PCR with Q5 High-Fidelity DNA Polymerase (New England Biolabs) with primers designed to amplify a 420-base-pair product around the variation point (forward: 5′-CTAGAAACCCCGAAACCAAA-3′, reverse: 5′-TCATACTAACAGTGTTGCATC-3′).
The PCR reaction was column-purified and analysed using Oxford Nanopore Technology sequencing (Plasmidsaurus). A custom Python script (available via GitHub at https://github.com/GreletLab/mtDNA-heteroplasmy) was developed specifically for this project to process the obtained raw reads, align them to the target sequence, classify each read as wild type (mouse host-derived) or mutated (cancer cell-derived) and generate statistical output. The script was validated using mtDNA sequence from pure 4T1 cell extract or BALB/c mouse tissue, as well as through the manual counting of some samples to ensure the accuracy of the output.
Intraductal human-in-mouse transplantation model
Intraductal transplantation of human ductal carcinoma in situ cells was performed as previously described13. In brief, before transplantation into 6-week-old virgin female SCID-beige mice, DCIS.COM cells were resuspended as single cells in PBS and counted. A 30-gauge Hamilton syringe, 50-μl capacity, with a blunt-ended 1/2-inch needle, was used to deliver the cells. The mice were anaesthetized, and a Y-incision was made on the abdomen to allow the skin covering the inguinal mammary fat pads to be peeled back to expose the inguinal gland. The nipple of the inguinal gland was snipped so that the needle could be directly inserted through the nipple. Two microlitres of cell culture medium (with 0.1% trypan blue) containing cells at a concentration of 2,500 to 5,000 cells per microlitre was injected; the injected liquid can be visually detected in the duct. The skin flaps were repositioned normally and held together with wound clips.
Chemical denervation
BoNT/A denervation was performed as previously described8, adapting the procedure through intraductal delivery of the toxin 4 weeks after the intraductal transplantation of the cells, and the experiment was concluded at 10 weeks. As previously described, the loss of nerve function, whether through physical or chemical denervation procedures, resulted in axonal atrophy and decreased intratumoural nerve density with an effect akin to Wallerian degeneration6,16,70,71. BoNT/A was reconstituted in 0.1 ml of 0.9% saline without preservatives to achieve a concentration of 1 U μl−1 and was utilized within 2 h of reconstitution. For the intraductal model, 15 U kg−1 of BoNT/A per xenograft was carefully injected using a 30-gauge needle and a precision glass syringe, with meticulous attention to avoid extra ductal leakage. For the mammary fat pad injection model, 0.3 units of BoNT/A in 100 µl PBS was injected into the mammary fat pad 1 week before the injection. One million 4T1 cancer cells (100 µl of a suspension of 10 million cells per ml) were injected into the same mammary fat pad. Using calipers, the tumour’s height and width were subsequently estimated.
Mammosphere assay
Cancer cells isolated from the MitoTRACER coculture and established as long-term subcultures were seeded at 5,000 cells per ml in 96-well ultralow-attachment plates. Mammospheres were collected on the seventh day of culture, washed with PBS, and sedimented for 30 min. All mammospheres with diameters greater than 50 µm were counted with a fluorescence microscope (BZ-X810, Keyence).
Invasion assay
Cell invasion was assessed using the Corning BioCoat Matrigel Invasion Chamber (Fisher Scientific), following the manufacturer’s instructions. In brief, the Matrigel-coated chambers were rehydrated with serum-free medium for 2 h at 37 °C in a humidified incubator with 5% CO2. After rehydration, 50,000 cells were seeded in the upper chamber in a serum-free DMEM medium; the lower chamber contained DMEM supplemented with 20% FBS as a chemoattractant. The plates were incubated for 24 h at 37 °C. Non-invaded cells and Matrigel were removed from the upper membrane surface using a cotton swab. The invaded cells on the bottom side of the membrane were fixed and stained with DAPI. Fluorescence microscopy was used to image the stained cells, and a custom image analysis script (available via GitHub at https://github.com/GreletLab/DAPI-count) quantified the number of cells that migrated through the membrane. The script was previously validated by traditional manual counting to compare to the output data and ensure its accuracy. All assays were performed in independent triplicate.
Shear stress assay
The shear stress assay was performed as previously described38. In brief, cancer cells were resuspended at a concentration of 1 × 105 cells per ml and loaded into the injection pump using a 30-ml syringe. The suspension was dispensed through a 26-gauge, 150-mm blunt tip Luer lock needle at a controlled flow rate of 0.25 ml s−1. The cells were subjected to 10 shear cycles, after which cell viability and quantity were assessed using a Celigo image cytometer. Cell viability was determined with Hoechst–propidium iodide staining to evaluate cell quantity and cell death.
Oxidative stress assay
To induce oxidative stress, cancer cells were cultured in vitro and treated with increasing concentrations of hydrogen peroxide. Cells were seeded at a density of 50 × 104 cells per well in a 96-well plate and allowed to adhere overnight. The following day, cells were exposed to a range of hydrogen peroxide concentrations from 100 mM to 9.375 µM in serum-free medium for 24 h. After treatment, cells were washed with PBS and analysed for recovery before Hoechst–propidium iodide staining to evaluate cell quantity and cell death using the Celigo image cytometer.
Calcium flux imaging
Nerve–cancer cocultures were grown in Nunc Lab-Tek II chambered coverglass slides (Roskilde) at 37 °C with 5% CO2. Cells were stained with Fluo-4AM calcium indicator dye and Hoechst 33342 (Invitrogen) just before imaging, following the manufacturer’s instructions. Time-lapse videos were acquired using a Nikon A1r confocal microscope equipped with a stage-top incubator for temperature and gas control, using a 20× objective lens (numerical aperture 0.8). Images were captured every 15 s over a duration of 10 min. Calcium fluorescence intensity was quantified by defining a region of interest around the cell to be measured and calculating the mean fluorescence intensity of the green channel within the region of interest for each time point.
Mouse xenograft, lineage tracing and fate-mapping experiments
Mixed-cell MitoTRACER spheroids were injected into the mammary fat pads of 6-week-old female BALB/c mice. In accordance with the Institutional Animal Care and Use Committee (IACUC) protocol for this study, tumour volumes were monitored every 2–3 days using digital calipers, and humane end points were applied if tumours exceeded 2 cm3 or signs of distress or ulceration appeared. Tumour volume was estimated through (short side × short side × long side)/2. The maximal size set by IACUC was 2 cm3 and was not exceeded in any experiment.
At end point, primary tumours, lungs and brains were collected, rinsed in PBS, and processed for dissociation, cell culture and flow cytometry to quantify red-to-green cell ratios in both primary and metastatic sites. Dissociated cell suspensions were filtered through 40-μm strainers, centrifuged at 300g for 5 min, and seeded in DMEM for further analysis. To confirm metastasis at distant sites, a subset of cells was cultured in vitro with Blasticidin selection for 2–4 weeks before re-examination of red and green fluorescence signals.
Tumour dissociation was performed using the Tumor Dissociation Kit (Miltenyi Biotec 130-096-730); tumours or organs weighing between 0.04 and 1 g were cut into 2–4-mm pieces and placed in 1–2.5 ml of enzyme medium. Tumours weighing more than 0.2 g were placed in C-tubes and processed using the gentleMACS Dissociator both before and after incubation. Samples not placed in C-tubes were manually separated after incubation using a filter. All samples were then strained through a 40-µm or 70-µm filter into a 50-ml tube and rinsed with 10–20 ml of RPMI or DMEM. The samples were centrifuged at 500g for 7 min, after which the supernatant was discarded, and flow buffer was added.
Dual immunohistochemistry and inForm analysis
The human breast cancer tissue microarray was purchased from Bio-Techne. The prostate cancer perineural array and controls were obtained at Baylor College of Medicine. Tissue sections from four patients with prostate cancer were obtained from the NCT01520441 phase 1/2 trial designed as a proof of principle that nerves affect the biology of cancer in humans. Patients served as their own controls by receiving BoNT/A injections into the right peripheral and transition zones and sham saline injections into the left peripheral and transition zones2. A paired treated cancer and control was available for only one patient and was used for this study.
Tissue microarray slides and clinical trial slides were dual-stained with antibodies to mitochondria (NeoBiotechnologies, catalogue number MSM2-740-P1ABX) and PGP 9.5 (Invitrogen, PA5-29012) using the Dako Omnis instrument with Envision Flex HRP and high-pH reagents (Agilent, Dako Omnis, GV800). Antigen retrieval was performed using EnV FLEX TRS at high pH (97 °C for 30 min). The PGP 9.5 antibody was used at a dilution of 1:1,500 with a 30-min room-temperature incubation, and the mitochondria antibody was used at 1:5,000 with a 30-min room-temperature incubation. Diaminobenzidine and magenta were used as chromogens for PGP 9.5 and mitochondria, respectively.
Slides with dual immunohistochemical staining were converted to high-resolution digital images through multispectral imaging using a Nuance Multispectral Microscope (PerkinElmer). The images were analysed using inForm analysis software (Akoya Biosciences), as previously described32. The system was trained to recognize and segment cancer, stroma and empty spaces. After segmentation of image compartments, each cell within the identified compartments was analysed separately for nuclear and cytoplasmic regions. Mitochondrial load was quantified by measuring the optical density of magenta staining specifically in the cytoplasm of cancer cells.
Statistics and reproducibility
Statistical analyses were conducted using Prism 10 for macOS (GraphPad) and Excel for MacOS (Microsoft). Unless otherwise specified in the figure legends, all in vitro data analyses were performed using unpaired t-tests for pairwise comparisons. For clinical data interpretation, high-throughput analysis of mitochondrial load in cancer cells from clinical histology samples was conducted. The equality of variances was first assessed using Levene’s test. A standard Student’s t-test was used for comparisons when variances were equal, and Welch’s t-test was applied when variances were unequal. For animal studies, unless specified otherwise, data were analysed using ANOVA with Tukey’s multiple comparison test. Two-tailed t-tests were used for exploratory experiments, and one-tailed t-tests were used when a directional effect was supported by prior evidence presented earlier in the study. The number of independent animals used is provided in the figure legends. Shear stress and Seahorse metabolic assays were conducted independently three times, with multiple culture replicates per assay. For Seahorse measurements, 8 to 16 technical replicates were included per assay, depending on the number of biological conditions tested in the 96-well plate. The experiments were analysed using the Wave Controller software (Agilent Technologies) and Prism 10 for MacOS, and graphs shown represent a typical experiment. Microscopy, western blot and PCR experiments were performed at least in triplicate, and representative images are provided. Uncropped western blots and PCR gels are provided in the Supplementary Information. P values are reported as exact values or symbolically as follows: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. A P value of less than 0.05 was considered to indicate nominal statistical significance.
Animals and housing
Six-week-old virgin female SCID-beige mice were used for the intraductal breast cancer model. The mammary fat pad injection model was performed on BALB/c female mice (age, 5 weeks) and flank injection of B16-F10 in C57BL/6 male mice (age, 6 weeks). Mice were housed under a 12 h/12 h light/dark cycle in a barrier vivarium. Room temperature (22.8 °C ± 1.7 °C) and relative humidity (30–70%) were continuously monitored and maintained. Animals were housed in cages of four, and each experimental condition was assigned to one cage. For each experiment, one cage was randomly selected from a pool of pre-assigned condition-specific cages. Although allocation was not randomized at the individual animal level, random selection at the cage level ensured unbiased assignment. All mice were age- and sex-matched, and housed under identical environmental and handling conditions to minimize potential covariates.
Institutional Review Board statement
The study was conducted according to the guidelines of the Declaration of Helsinki. All experiments and procedures were conducted in accordance with the guidelines described in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). Approvals for animal work were obtained from the Baylor College of Medicine Animal Care and Use and Human Subjects Committee and the University of South Alabama IACUC. Informed consent was obtained from all human participants involved in the study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.