Mouse studies
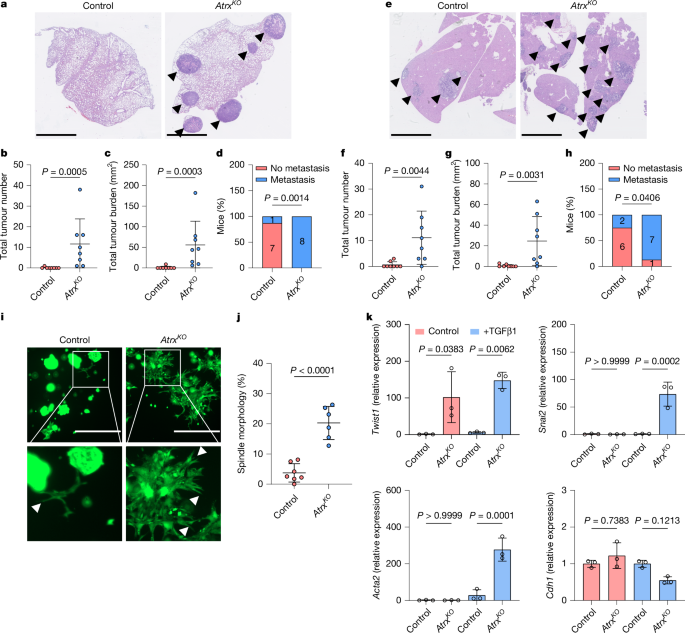
All in vivo experiments were performed in accordance with the UK Home Office regulations (project licence: PP9016178, PP7510272 and PP3908577) and were subject to local ethics review by the animal welfare and ethics board of the University of Edinburgh and University of Glasgow. Mice were housed under a 12-h light–dark cycle at a temperature ranging from 19 to 23 °C. Ambient humidity was 55 ± 10%. Standard diet and water were available ad libitum. Work was performed on the C57BL/6J and the CD1 nude background available from Charles River Laboratories. For the lung metastatic model, 500,000 AKP control or AKP AtrxKO cells were resuspended in 100 µl PBS and injected into the tail vein of female CD1 nude mice. For the liver metastatic model, C57BL/6J mice were anaesthetized with isoflurane and splenic access was achieved using laparotomy. Around 500,000 cells were injected in 50 µl PBS, following which, the incision was closed with staples. Subcutaneous injections into female CD1 nude mice were performed with 50,000 AKP control, AKP AtrxKO or AKP Hnf4aKO cells resuspended in 100 µl basement membrane extract (BME). Subcutaneous injections into female CD1 nude mice were performed with 12,000 flow cytometry-sorted cells resuspended in 100 µl BME.
Colonic submucosal injections were performed using a Karl Storz TELE PACK VET X LED endoscopic video unit. AKP control, AKP AtrxKO, BPN control and BPN AtrxKO organoids were dissociated by mechanical pipetting and then were washed with PBS before being injected orthotopically in female CD1 nude mice. Approximately 500 organoids in 70 μl PBS were injected in a single injection. The BPN organoid line (male) was generated in the laboratory of O.J.S.
In accordance with the respective licence and study protocol, mice were humanely euthanized at the reported experimental end points (15 mm diameter for subcutaneous injection) as defined in the relevant licensing documents.
In accordance with the 3Rs (replacement, reduction and refinement), the smallest sample size was chosen that could give a significant difference.
C57BL/6J or CD1 nude mice 6–12 weeks of age were randomly grouped for transplantation experiments. All mice received the same number of cells of a different genotype or phenotype. Experimental groups were determined by the genotype of injected cells (for example, AKP control versus AKP AtrxKO). Investigators were blinded to the genotype of tumours when monitoring for clinical signs, when carrying out histological analyses and during data collection. IHC analysis of tumour histology was carried out using QuPath software, with the investigator blinded to the tumour genotype.
Histopathology, IHC and immunofluorescence
Tissues were fixed overnight in formaldehyde 4% stabilized, buffered (VWR) and then transferred to 70% ethanol. Tissue processing was done using a Tissue-TeK VIP infiltration processor (Sakura) followed by embedding in paraffin wax. Tissues were sectioned at 5 µm thickness, dried at 37 °C and then deparaffinized and rehydrated. For each sample, one section was stained with haematoxylin and eosin. Additional sections were used for either standard IHC or immunofluorescence analyses. The following primary antibodies were used: KRT5 (rabbit; Abcam 52635; 1:200 or chicken; BioLegend 905903; 1/200); LY6D (rabbit; Atlas HPA024755; 1:200); TWIST (mouse; Santa Cruz 81417, 1:200); EPCAM (rabbit; Abcam 71916; 1:200); HNF4A (rabbit; CST 3113; 1:500); ATRX (mouse; Sigma MABE1798; 1:500); β-catenin (mouse; BD 610154; 1:50); E-cadherin (rabbit; CST 3195, 1:200); and CDX2 (mouse; Atlas AMAb 91828, 1:1,000). IHC secondary detection was achieved using EnVision+ System-HRP labelled polymer (Dako). Positive signals were visualized using DAB substrate (Epredia and 2b scientific) for 5–10 min. Sections were counterstained with haematoxylin. For immunofluorescence studies, the following secondary antibodies (1:400) were used: anti-rabbit-594 (Invitrogen, A21207); anti-chicken-488 (Invitrogen, A78948); anti-streptavidin-647 (Invitrogen, S32357); and anti-rabbit-488 (Abcam, 150073) Representative images are shown for each stain. The slides were digitized using a Nanozoomer digital slide scanner (Hamamatsu) with NDP.view2plus software and analysed using QuPATH (v.0.2.3). The commercial colon cancer human tissue array used was CO804b (Biomax) (Extended Data Fig. 1b–d). The TMA in Fig. 5 and Extended Data Fig. 10 includes patients with stage I–III CRC who underwent potentially curative resection between 1997 and 2013 at Glasgow Royal Infirmary. Three cores were taken from each donor block at the invasive edge, capturing a small amount of tumour and the environment around it. Patient tissue access was authorized by the NHS Greater Glasgow and Clyde Biorepository under their NHS Research Ethics Committee, with ethics approval granted in biorepository application number 845, West of Scotland Ethics 22/WS/0207 in accordance with recognized ethical guidelines as described in the Declaration of Helsinki. Overall, 17 patients undergoing synchronous resection of primary CRC and CRC liver metastases with curative intent between April 2002 and June 2010 at Glasgow Royal Infirmary were analysed. Patient tissue access was authorized by the NHS Greater Glasgow and Clyde Biorepository under their NHS Research Ethics Committee, with ethical approval granted in biorepository application number 357, West of Scotland Ethics 22/WS/0207 in accordance with recognized ethical guidelines as described in the Declaration of Helsinki.
H-scores were generated for HNF4A, ATRX and CDX2 tumour cell expression using QuPath-0.2.3. Tumour cores with >2% tumour cells positive for LY6D and KRT5 expression were designated as LY6D+ and KRT5+, respectively. Tumour cores with <2% tumour cells positive for LY6D or KRT5 expression were designated as LY6D– and KRT5–, respectively. Analysis was performed using QuPath-0.2.3. H-scores were generated for CDH1 cell expression using QuPath-0.2.3.
Mouse organoid studies
Organoids were resuspended in Cultrex reduced growth factor BME type 2 (Bio-Techne), plated in a 10 µl drop on a culture plate and cultured in organoid culture medium containing advanced DMEM–F12 (Gibco) medium supplemented with 100 units ml–1 penicillin, 100 µg ml–1 streptomycin, 2 mM l-glutamine, 10 mM HEPES (all from Life Technologies), 1 ml Primocin (Invivogen), N2, B27 (both from Gibco), 50 ng ml–1 EGF (Peprotech) and 1% Noggin conditioned medium. The Noggin-producing cell line was a gift from H. Clevers’ group (Hubrecht Institute). Organoids were passaged by mechanical fragmentation with a p1000 and p200 pipette. All organoids were grown in a humidified incubator at 37 °C supplemented with 5% CO2. Single cells were generated by incubating organoids in TrypLE Express (Life Technologies) for 15 min at 37 °C and passed through a 40 µm strainer. Cell counting was performed using a Countess II automated cell counter (Invitrogen). To study the effect of TGFβ, dissociated cells (10,000 (or 5,000 for the Tgfbr2KO) cells in 10 µl BME) were cultured in organoid culture medium in the presence of 5 ng ml–1 TGFβ (Peprotech). TGFβ treatment in Extended Data Fig. 3 was performed using a different range of concentrations as specified in the figure. On day 3 the medium was replaced with fresh organoid culture medium containing TGFβ. Cells were collected on day 13 for RNA extraction. Alternatively, cells were stained with calcein (Abcam) or phalloidin-568 (VWR) within 14 days of treatment.
For ITGA5+ cell sorting and TGFβ treatment, AKP control and AKP AtrxKO organoids were digested as described above, and ITGA5 FACS was carried out as described in the FACS section below. In brief, 10,000 ITGA5+ or ITGA5– cells were plated in 10 µl BME and treated with 5 ng ml–1 TGFβ as described above. On day 13, organoids were fixed with 4% paraformaldehyde, stained with phalloidin-568 and analysed for the presence of spindle-like structures.
For the tail-vein injections, organoids were dissociated in TrypLE Express (Life Technologies) supplemented with a Rho kinase inhibitor (Y-27632, Tocris) for 15 min at 37 °C, passed through a 40 µm strainer, counted and resuspended in PBS. For splenic injections, organoids were collected, washed in PBS, mechanically fragmented by vigorous pipetting and then incubated for 7 min at 37 °C in 0.25% trypsin in EDTA–PBS. After quenching of trypsinization by immersion in 10% FBS, cells were passed through a 40 µm strainer, counted using a haemocytometer and resuspended in PBS to achieve a final volume of 1 × 107cells per ml. Cells were routinely tested for mycoplasma contamination.
Human patient-derived organoids
The patient-derived organoids used in this study were generated by F.V.N.D. and M.G.D. MD20043 is an 81-year-old man with stage 4 rectal cancer: T3, N2, M1 (where ‘T’ is tumour, ‘N’ is nodes and ‘M’ is metastases). Ethics approval for human CRC organoid derivation was carried out under NHS Lothian Ethical Approval Scottish Colorectal Cancer Genetic Susceptibility Study 3 (SOCCS3) (REC reference: 11/SS/0109). The patient provided fully informed consent for the use of their tissues.
Human organoid culture medium and ATRX KO generation
Human carcinoma organoids were cultured in advanced DMEM–F12 (Gibco) medium supplemented with 100 units ml–1 penicillin, 100 µg ml–1 streptomycin, 2 mM l-glutamine, 10 mM HEPES (all from Life Technologies), 1 ml Primocin (Invivogen), 1% Noggin conditioned medium (The Noggin-producing cell line was a gift from H. Clevers’ group, Hubrecht Institute), B27 (Gibco), 50 ng ml–1 EGF (Peprotech), 10 nM gastrin (Sigma), 10 nM PGE2 (Tocris), 10 mM nicotinamide (Sigma), 10 µM SB202190 (Sigma), 600 nM A83-01 (Biotechne) and 12.5 mM N-acetylcysteine (Sigma).
For transduction conditions, human organoids were pretreated with IntestiCult (Stem Cell Tech) organoid growth medium and 1 mM valproic acid (Merk) for 48 h after 2 days post-split (day 1). On transduction day (day 1), human organoids were digested into a single-cell suspension in TrypLE Express (Life Technologies) with 10 µM Y-27632 (Tocris) for 8 min at 37 °C with mechanical dissociation every 4 min. Single-cell suspensions were combined with viral particles containing a non-targeting or ATRXKO sgRNA (hATRX: 5′-GCTATAAACAGAAAAAGAAA-3′; pLentiV2-Addgene) and placed on a BME layer. On day 2, medium was changed into IntestiCult supplemented with 1 mM valproic acid and 10 µM Y-27632. On day 3, antibiotic selection started with IntestiCult supplemented with 10 µg ml–1 blasticidin (Gibco) for 3 weeks. IntestiCult medium was used exclusively during transduction and the single-cell stage of organoid development for clone selection. Otherwise, the above-described medium was used for maintaining the organoids.
In vitro drug treatment and cell-proliferation assay
Organoids were dissociated in TrypLE Express (Life Technologies) for 15 min at 37 °C, passed through a 40 µm strainer and resuspended in BME. Next, 1,000 single cells in 10 µl BME were plated in 24-well plates and treated with 250 nM JQ1 (Stratech), 0.25 ng ml–1 IFNγ (Thermo Fisher Scientific), 10 nM FK228 (Stratech) or (0.5 ng ml–1) TNF (Peprotech) for 7 days.
For Resazurin cell viability assays, Resazurin (R&D systems) was added at a volume equal to 10% of the cell culture medium volume and incubated at 37 °C. Fluorescence was read using 544 nm excitation and 590 nm emission wavelengths.
RNA extraction, RT–qPCR and RNA-seq
Total RNA was isolated using a RNeasy Mini kit (Qiagen) accordingly to the manufacture’s protocol. RNA was then subjected to DNA-free DNase treatment (Invitrogen). cDNA was generated using 1 µg RNA by reverse transcription using qScript cDNA SuperMix (Quantabio). RT–qPCR was performed using SYBR Select master mix (Applied Biosystems). Ct values were normalized to β-actin or 18S rRNA. The ΔΔCt method was used to calculate relative gene expression values. Oligonucleotides used in this study are listed in Supplementary Table 11. For the RNA-seq experiments, RNA integrity was evaluated using an Agilent 2200 Bioanalyser. Truseq mRNA-seq libraries were prepared from total AKP and AKP ATRXKO RNA, and these were then sequenced using NovaseqS1 Illumina sequencing at Edinburgh Genomics Facility. AKP and AKP Hnf4aKO libraries were prepared from 100 ng of each total RNA sample using a NEBNEXT Ultra II Directional RNA Library Prep kit (NEB 7760) and the Poly-A mRNA magnetic isolation module (NEB E7490) according to the provided protocol. Sequencing was performed on a NextSeq 2000 platform (Illumina, 20038897) using NextSeq 2000 P3 reagents (200 cycles) (20040560). RNA-seq analysis was carried out using the RaNA-seq pipeline with default settings32.
Western blotting
Cells were lysed using RIPA buffer (Sigma) supplemented with 1% phosphatase and protease inhibitors (Sigma). Protein concentration was measured using a BCA Protein Assay kit (Pierce). A total of 20 µg protein lysate was resuspended in 4× LDS sample buffer (Invitrogen) supplemented with sample reducing agent (Invitrogen) and denatured at 100 °C for 5 min. Proteins were separated by electrophoresis on NuPAGE 3–8% Tris-acetate protein gels (Invitrogen) using Tris-acetate buffer and blotted onto an activated PVDF or nitrocellulose (Cytiva) membrane at 100 V for 1.15 h. Membranes were incubated in blocking solution (5% milk, 0.1% Tween-20–PBS) for 1 h at room temperature, and then in primary antibody. The following primary antibodies were used: β-actin (Cell Signalling Technology, 1:5,000); ATRX (MABE1798, Sigma; 1:500); and HNF4A (C11F12, Cell Signalling Technology, 1:,1,000). After 3× 10-min washes in 0.1% Tween-20–PBS, the membrane was incubated in HRP-linked secondary antibody for 1 h at room temperature. The following secondary antibodies were used: anti-rabbit or anti-mouse IgG HRP-linked (Cell Signalling Technology, 1:5,000). Following 3× 10-min washes in 0.1% Tween-20–PBS, antibody signals were detected by using ECL Plus Western blotting substrate (Pierce) and visualized using an ImageQuant 800 (GE Healthcare). Full scans are provided in Supplementary Information 1 and 2.
CRISPR–Cas9 genome editing
sgRNAs (mAtrx: 5′-ACGGCGCATTAAGGTTCAAG-3′; mHnf4a 5′-CGGGCCACCGGCAAACACTA-3′, mTgfbr2: 5′-AAGCCGCATGAAGTCTGCG-3′, non-targeting controls 5′- GCTTTCACGGAGGTTCGACG-3′ or 5′-ATGTTGCAGTTCGGCTCGAT-3′) were cloned individually into lentiCRISPR v.2 plasmids (Addgene) following Addgene’s protocol. Lentiviral particles were generated using HEK293T cells (provided by J. C. Acosta (IGMM, Edinburgh), originally obtained from the American Type Culture Collection): 10 µg gene-specific lentiviral vector was mixed with 7.5 µg lentiviral packaging vector psPAX2 and 2.5 µg envelope-protein-producing vector pCMV-VSV-G (both from Addgene) and transfected into HEK293T cells in a 10 cm2 dish using polyethylenimine as the transfection reagent (Polysciences). After 48 h, the supernatant medium was filtered using a 0.45 µm syringe filter and concentrated using a Lenti-X concentrator (Takara Bio). Lentiviral transduction was carried out as previously described33. In brief, AKP organoids were expanded and cultured in organoid culture medium supplemented with 10 µM Rho kinase inhibitor (Y-27632, Tocris) and 1 mM valproic acid (Sigma) for 48 h. Spheroids were enzymatically dissociated with StemPro Accutase cell dissociation reagent (Gibco) supplemented with 10 µM Y-27632 for 3 min at 37 °C. Dissociated organoids were then washed twice with advanced DMEM–F12 medium. Cells were counted using a Countess II Automated cell counter (Invitrogen). Next, 5 × 105 cells were plated on a 150 µl bed of BME in a 6-well plate in the presence of Y-27632, 1 mM valproic acid and 4 µg ml–1 polybrene (Sigma). Virus was removed 24 h after transduction, and adhered organoids were overlaid with 150 µl BME. To select transduced cells, 2 µg ml–1 of puromycin or 10 µg ml–1 blasticidin (both from Gibco) was added to the organoid culture medium supplemented with 10 µM Y-27632. Multiple deleted clones were generated. Editing of clonal lines was confirmed by genomic sequencing or western blotting.
TissueEnrich analysis
TissueEnrich analysis34 (https://tissueenrich.gdcb.iastate.edu/) was carried out using the web-based tool with the following settings: gene symbol, Homo sapiens, Human Protein Atlas, All. Lists of gene upregulated or downregulated by >2 fold in the AKP versus AKP AtrxKO dataset were used for input. In Fig. 2g, TissueEnrich analysis was performed on the list of genes upregulated in scRNA-seq cluster 4 and cluster 15 (fold change > 1.5).
ATRX mutation analysis in CRC transcriptional subtypes
For ATRX mutation enrichment analysis, we used previously designated CRIS subtypes14 and analysed TCGA mutational data for ATRX mutations for which the CRIS designation is known. The number of tumours carrying ATRX mutations was calculated for the CRIS-B subtype and compared with all other subtypes using Fishers’ exact tests. For survival analysis, only CRIS-B tumours were analysed using TCGA survival data downloaded from the TCGA data portal. For iCMS, the same analysis was carried out but with TCGA data separated on the basis of iCMS designation.
HNF4A overexpression
HNF4A overexpression was generated using the pGCDNsam-HNF4A-IRES-GFP plasmid (Addgene). In brief, AKP Atrx organoids were transduced as above. To select transduced cells, FACS analysis for GFP positivity was performed.
Collection of human samples and processing for FACS
Normal colorectal mucosa and tumour were sampled from freshly resected surgical specimens from patients diagnosed with CRC. Ethics approval was carried out under NHS Lothian Ethical Approval Scottish Colorectal Cancer Genetic Susceptibility Study 3 (SOCCS3 REC: 11/SS/0109, IRAS: 9556). All patients provided fully informed consent for the use of their tissues. Tissues were cut into small pieces and then incubated in Advanced DMEM–F12 supplemented with 1 mg ml–1 collagenase type IV (Sigma), 0.5 mg ml–1 hyaluronidase (Sigma) and 10 µM Y-27632 (Tocris) at 37 °C with vigorous shaking until the tissue was completely disaggregated (60–90 min). The digested reaction was then filtered through a 70 µm cell strainer. The filtered cells were centrifuged at 500g for 5 min, washed twice in Advanced DMEM–F12 and once in 0.1% BSA in PBS. Single-cell suspensions were then analysed by FACS.
FACS
Pelleted organoids were resuspended in 1 ml TrypLE Express (Gibco) and incubated at 37 °C for 15 min. Cells were vigorously dissociated by pipetting, resuspended in 10 ml advanced DMEM–F12, passed through a 40 µm cell strainer and centrifuged at 300g for 5 min at 4 °C. Single cells were washed with 0.1% BSA in PBS and stained with the following antibodies: EPCAM–APC (BioLegend, 118213; 1:200); LY6D–PE (BioLegend, 138603; 1:200) or LY6D–APC (Miltenyi, 130-115-313; 1:50); and ITGA5–PE (BioLegend, 103805; 1:200). Human single-cell suspensions were stained with EPCAM–APC (BioLegend, 324207; 1:50) and LY6D–FITC (Cusabio Biotech, CSB-PA613492LC01HU; 1:50). Cells were then washed twice in 0.1% BSA in PBS before being subjected to FACS (BD FACSARIA II/BD LSR-Fortessa X-20). Single viable cells were gated by negative staining for DAPI. The gating strategy is provided in Supplementary Information 3. Analyses were performed using FlowJo (v.10.8) software.
scRNA-seq data processing
Raw sequencing reads were processed and aligned to the mouse reference genome (mm10) using the 10x Genomics CellRanger pipeline (v.7.2.0). The gene expression matrices obtained from CellRanger were analysed using the R package Seurat (v.5). Cell barcodes with <200 unique genes and >10% mitochondrial gene expression were removed. The filtered matrices were normalized to the total unique molecular identifier counts per cell, and cell cycle effects were regressed out. Data integration was performed with Harmony, and the subsequent Louvain clustering resulted in 18 clusters. Differentially expressed genes between clusters were obtained using Wilcoxon rank-sum tests implemented by the FindAllMarkers function in Seurat.
ATAC–seq
ATAC–seq was performed using an Active Motif commercially available kit as per the manufacturer’s instructions. In brief, 50,000 cells were lysed with ATAC lysis using a pestle and dounced slowly for 25 strokes on ice followed by Tn5 tagmentation for 30 min at 37 °C. Size selection was performed using the SPRIselect protocol (Beckam). After indexing and PCR amplification, DNA libraries were multiplexed and sequenced with an Illumina Nextseq 2000 on a 100-cycles kit by Edinburgh Clinical Research Facility–Wellcome Trust CRF.
ATAC–seq analysis
Raw sequencing output fastq files were inputed into an nf-core ATAC–seq pipeline using default pipeline settings (https://nf-co.re/atacseq/2.1.2)35. ATAC–seq library quality was assessed for the presence of contaminating mitochondrial DNA sequences. Pipeline output bigWig files were used for creating locus-specific genome images from the IGV browser (https://www.igv.org/). ATAC–seq peaks were analysed for differential enrichment between samples using DESeq2 (ref. 36). Principal component analysis was performed on tables of ATAC-seq read counts (DESeq2 output) using base R scripts. Subsequently, DESeq2 ATAC–seq read counts were input into the monaLisa R package37 to determine the presence of differentially enriched TF-binding or DNA-binding site motifs between sample groups.
CUT&RUN
Cells were digested with TrypLE Express at 37 °C for 15 min. Next, 1 × 106 viable cells were fixed in PBS containing 0.1% formaldehyde at room temperature for 1 min. Formaldehyde was quenched with glycine at 0.125 M, followed by 5 min of incubation at room temperature. Fixed cells were pelleted and washed twice with room temperature wash buffer (20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine (Sigma-Aldrich), 1% Triton X-100, 0.05% SDS and 1× protease inhibitor cocktail (Roche)). Cells were pelleted and resuspended in 100 µl wash buffer. The cell suspension was added at a 10:1 ratio to concanavalin A beads (CST) activated as per the manufacturer’s instructions, gently mixed and incubated at room temperature for 20 min. The cell–bead slurry was placed on a magnetic rack until clear, the supernatant removed and samples resuspended in cold antibody buffer (20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine, 0.0025% digitonin (Thermo Fisher), 2 mM EDTA, 1% Triton X-100, 0.05% SDS and 1× protease inhibitor cocktail (Roche)). The following antibodies were added and incubated at 4 °C overnight: XP isotype control DA1E (rabbit; CST 1:10) or anti-histone H3, acetyl K27 (rabbit; Abcam 1:250). Samples were washed twice with digitonin buffer (20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine, 0.0025% digitonin, 1% Triton X-100, 0.05% SDS and 1× protease inhibitor cocktail (Roche)) and resuspended in digitonin buffer. 1× CUTANA pAG-MNase (EpiCypher) was added and incubated for 10 min at room temperature. Samples were washed twice with digitonin buffer. Chromatin was cleaved by the addition of 2 mM CaCl2 and incubation for 2 h at 4 °C. Digestion was halted by the addition of stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA and 50 µg ml–1 glycogen (ThermoFisher)) and chromatin released by incubation at 37 °C for 10 min. The supernatant was collected and SDS added to a final concentration of 0.09%. Proteinase K (CST) was added and incubated overnight at 55 °C. DNA was purified using MaXtract High Density columns (Qiagen) as per the manufacturer’s instructions and precipitated with GlycoBlue (ThermoFisher) as per the manufacturer’s instructions. DNA was eluted in TE buffer (ThermoFisher). Samples were quantified and assessed for size distribution using an Agilent 2100 Electrophoresis Bioanalyser Instrument with a DNA HS kit (Agilent Technologies). Libraries were generated using a Simple-ChIP DNA library prep kit (CST) and quantified by fluorometry using a Qubit dsDNA HS assay and assessed for size distribution on an Agilent Bioanalyser with a DNA HS kit. Next, 100 bp paired-end sequencing was performed on a NextSeq 2000 platform (Illumina) using a NextSeq 1000/2000 P1 reagents (300 cycles) kit.
CUT&RUN data analysis
The nf-core CUT&RUN analysis pipeline (https://doi.org/10.5281/zenodo.7715959) was used to process CUT&RUN data using default parameters. Reads were normalized to spike in DNA (CST) and aligned to the mm10 reference genome. Regions in the mm10 ENCODE blacklist were removed38. MACS2 (ref. 39) was used for peak calling and peak calls were normalized to IgG controls. Differential analysis was conducted using DiffBind40 with default parameters. BEDTools41 was used to identify overlapping regions of significant difference in the H3K27ac CUT&RUN with the ATAC dataset.
TCGA patient stratification based on ATRX transcriptional signatures
Bulk RNA transcripts per kilobase million values for patients with colorectal adenocarcinoma from TCGA27 were obtained from the Genomic Data Commons Portal and were log2 normalized. Patients who had ATAC–seq data (n = 36) were stratified into high, medium or low for AtrxKO and AtrxWT signatures. This was performed by calculating the single-sample GSEA score using GSVA in R and selecting the samples above the third, second or first quantile for the respective category. These patients were also assigned an iCMS class as previously described23.
Differential analysis of TCGA bulk ATAC–seq data
Normalized ATAC–seq peak counts were obtained from the Genomic Data Commons Portal for 77 samples from 36 patients with colorectal adenocarcinoma; details on the normalization can be found the original publication27. In brief, the peak counts matrix was normalized using ‘cpm(matrix, log = TRUE, prior.count = 5)’ in edgeR followed by quantile normalization using normalize.quantiles of preprocessCore in R. A SummarizedExperiment object was created with the normalized count matrix and metadata for the patients. A two-tailed t-test was used to identify the peaks that have a significantly different mean count between the samples from patients categorized as HiSquam (high for AtrxKO and low for AtrxWT) or HiCol (low for AtrxKO and high for AtrxWT). The selection of differential peaks was based on FDR < 0.01 and Δlog2counts > 1 cut-off values. Counts from all samples for the set of significantly differential peaks were plotted into a heatmap using ComplexHeatmap in R.
HOMER TF motif enrichment
TF motif enrichment analysis was performed on the set of peaks that were differentially accessible for patients categorized as HiSquam or HiCol by using HOMER. The peaks were first annotated using ChIPeakAnno and then formatted into HOMER input style. The analysis was performed with the command findMotifsGenome.pl, genome “hg38” and “-size 200 -mask” as options. TF motif enrichment was presented as previously described42.
Analysis of bulk gene expression data with Atrx
WT and Atrx
KO signature genes
Normalized gene expression data of samples from patients with CRC were obtained from the Gene Expression Omnibus (accession GSE39582), and 557 patients with relapse survival information were used for the downstream analysis. HiCol, intermediate and HiSquam groups were identified by hierarchical clustering using AtrxWT and AtrxKO signature genes. The relapse-free survival of three patient groups was determined by Kaplan–Meier survival with log-rank tests.
Identification of Atrx
WT and Atrx
KO signatures from single-cell resolution
To understand the association between ATRXKO-driven gene expression and iCMS classification at the single-cell level, we performed the analysis using scRNA-seq data after downloading from syn26844071 (ref. 23) and used tumour cells with iCMS classification from primary tumour tissue samples after excluding patients with low numbers of cells. The dimension reduction analysis was performed using 715 iCMS-associating genes, and patient batch was corrected using Harmony43. AtrxWT and AtrxKO scores for each cell were calculated using the average expression level of each signature and then subtracted with the aggregated expression of the control gene score sets. The control gene score was calculated using the average expression of 100 randomly selected genes, replicated 10 times.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9. Statistical tests used are indicated in figure legends and exact P values shown throughout.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.