Reagents
The following chemicals and reagents were used: Dounce homogenizer (DWK Life Sciences, 885302-0002); Pierce anti-HA magnetic beads (Thermo Scientific, 88837); Pierce anti-Flag magnetic agarose (Thermo Scientific, A36797); anti-Flag M2 magnetic beads (Sigma Millipore, M8823); Pierce protein A/G magnetic beads (Thermo Scientific, 88802); IGEPAL CA-630 (Sigma-Aldrich, I8896); S-Trap micro columns (Protifi, C02-micro-80); triethylammonium bicarbonate (TEAB) buffer (Sigma-Aldrich, T7408); sodium dodecyl sulfate (SDS; Bio-Rad, 1610302); DSSO (Thermo Scientific, A33545); DHSO (CF Plus Chemicals, PCL042); DMTMM (Sigma-Aldrich, 74104); n-dodecyl β-d-maltoside (DDM, Gold Biotechnologies, DDM5); NativeMark protein standard (Invitrogen, LC0725); NativePAGE 4–6% gels (Invitrogen, BN1002BOX); MultiScreen filter plates (Sigma Millipore, MSHVN4510); TMTpro 16plex set (Thermo Fisher Scientific, A44520); protease inhibitor cocktail (Roche, 4906845001); tris(2-carboxyethyl)phosphine (TCEP; Gold Biotechnology, 51805-45-9); 2-chloroacetamide (Sigma-Aldrich, C0267); S-methyl thiomethanesulfonate (MMTS; Sigma-Aldrich, 208795); trypsin (Promega, V511C); Lys-C (Wako Chemicals, 129-02541); hydroxylamine solution (Sigma-Aldrich, 438227); Sep-Pak C18 and C8 50 mg cartridge (Waters, WAT054955 and WAT054965); high-pH reversed-phase peptide fractionation kit (Thermo Scientific, 84868); Bio-Rad protein assay dye (Bio-Rad, 5000006); 3-[4-(2-hydroxyethyl)-1-piperazine]propanesulfonic acid (Thermo Scientific, J61296AE); Empore SPE discs C18 (Sigma Millipore, 66883-U); Gateway LR Clonase II enzyme mix (Thermo Scientific, 11791020); NEBNext Ultra II Q5 Master Mix (New England BioLabs, M0544L); Cas9-NLS (QB3 MacroLab at University of California, Berkeley); CloneR (StemCell Technologies, 05889); MiSeq reagent nano kit v2 (300 cycles; Illumina, MS-103-1001); GeneArt Precision gRNA synthesis kit (Thermo Fisher Scientific, A29377); RNAeasy Qiagen kit (Qiagen, 74104); 24-well glass-bottom plates (Cellvis, P24-1.5H-N); Corning square culture dish (Corning, 431110); Nunc Nunclon Delta cell culture dishes (Thermo Scientific, 140675, 150318 and 168381); Corning Matrigel matrix (Corning, 354230); DMEM with F-12 (Gibco, 11330057); neurobasal medium (Thermo Scientific, 21103049); non-essential amino acids (Gibco, 11140050); GlutaMAX (Gibco, 35050061); N-2 supplement (Gibco, 17502048); neurotrophin-3 (NT3; Peprotech, 450-03); brain-derived neurotrophic factor (BDNF; Peprotech, 450-02); B27 (Gibco, 17504001); Y27632 dihydrochloride (ROCK inhibitor; PeproTech, 1293823); Cultrex 3D culture matrix laminin I (R&D Systems, 3446-005-01); accutase (StemCell Technologies, 07922); FGF2-G3 (in-house); human insulin (Santa Cruz Biotechnologies, sc-360248); transforming growth factor-β (PeproTech, 100-21); holo-transferrin human (Sigma-Aldrich, T0665); sodium bicarbonate (Sigma-Aldrich, S5761-500G); sodium selenite (Sigma-Aldrich, S5261-10G); doxycycline (Clontech Labs, 631311); UltraPure 0.5 M EDTA (Invitrogen, 15575020); 16% paraformaldehyde (Electron Microscopy Science, 15710); DMEM (Gibco, 11995073); fetal bovine serum (Cytiva, SH30910.03); hydrocortisone (Sigma-Aldrich, H0135); polyethylenimine (Polysciences, 23966); FuGENE (Promega, E2311).
The following primary antibodies were used (1:1,000 for immunoblotting, 1:400 for immunofluorescence): Flag (Sigma-Aldrich, F1804), HA (Cell Signaling Technology, 3724), V5 (Invitrogen, 14-6796-82), TMEM230 (Origene, TA504888), LAMP1 (Cell Signaling Technology, D2D11), RAB5 (Cell Signaling Technology, C8B1), CLR (ProteinTech, 10292-1-AP), golgin 97 (ProteinTech, 12640-1-AP), VDAC1 (ProteinTech, 55259-1-AP), CLCN3 (Cell Signaling Technology, 13359S), GFP (Thermo Scientific, a10262), mCh (Thermo Scientific, M11217), EEA1 (Cell Signaling Technology, C45B10). The following secondary antibodies were used (1:10,000 for immunoblotting, 1:400 for immunofluorescence): anti-rabbit immunoglobulin-G (IgG) horse radish peroxidase (HRP) conjugate (Bio-Rad, 1706515); anti-mouse IgG HRP conjugate (Bio-Rad, 1706516); goat anti-chicken IgY (H + L), Alexa Fluor 488 (Thermo Scientific, A-11039); goat anti-rat IgG (H + L) cross-adsorbed, Alexa Fluor 555 (Thermo Scientific, A-21434); goat anti-rabbit IgG (H + L) cross-adsorbed, Alexa Fluor 647 (Thermo Scientific, A-21244).
Molecular cloning
Plasmids were made as previously described61. Entry clones from the human ORFeome collection, version 8, were cloned into their corresponding plasmids using Gateway technology (Thermo Fisher Scientific) or Gibson assembly (New England Biolabs). The complete TMEM230(Y29C/R78L/X121W) mutant was obtained by gene synthesis (Twist Bioscience). For lentivirus transduction, pHAGE and pLenti backbones were used. For transfection, pCGS and pcDNA3.1 backbones were used. The following plasmids were generated: pGCS-3×Flag-ATP11B (Addgene, 225511), pcDNA-TMEM30A-V5 (Addgene, 225510), pGCS-3×HA-TMEM230 (Addgene, 225512), pGCS-3×HA-TMEM230(Y29C/R78L/X121W) (Addgene 225513), pLenti-UBC-HA-TMEM230 (Addgene, 225516), pLenti-UBC-HA-TMEM230(R78L) (Addgene, 225517), pLenti-UBC-HA-TMEM230(X121W) (Addgene, 225519), pLenti-UBC-HA-TMEM230(Y29C) (Addgene, 225520), pLenti-UBC-HA-TMEM230(X121PG) (Addgene, 225518), pLenti-UBC-HA-TMEM230(Y29C/R78L/X121W) (Addgene 225521), pcDNA-CLCN3-3×Flag (Addgene, 225506), pcDNA-CLCN5-3×Flag (Addgene, 225507), pcDNA-TMEM9B-3×HA (Addgene, 225509), pcDNA-TMEM9-3×HA (Addgene, 225508), pHAGE-mCh-CLCN3 (Addgene, 225514), pHAGE-TMEM9-EGFP (Addgene, 225515). The following plasmids were used for lentiviral packaging: pPAX2 (Addgene, 12259), pMD2 (Addgene, 12260).
Cell culture, neuronal differentiation and lentiviral transduction
HEK293 cells (ATCC; RRID:CVCL_0045) were cultured in 10-cm dishes with high-glucose and pyruvate DMEM supplemented with 10% fetal bovine serum. For co-IP experiments, cells were transfected at 60% confluency with 6 μg of plasmids in a 2:1 ratio using polyethylenimine (25 kDa) and incubated for 48 h at 37 °C and 5% CO2. SUM159PT cells (a gift from T. Walter, Memorial Sloan Kettering; RRID:CVCL_5423) were cultured in 6-well culture dishes (300,000 cells per well) in DMEM with F-12 supplemented with GlutaMAX, 5% fetal bovine serum, 1 μg ml−1 hydrocortisone and 5 μg ml−1 insulin. Cells were transfected 1 day later with 500 ng of plasmids using FuGENE and Optimem transfection reagent and incubated at 37 °C and 5% CO2. One day after transfection, cells were selected with puromycin and plated into 24-well glass-bottom culture dishes (50,000–100,000 cells per well).
Gene-edited human embryonic stem (ES) cells (H9, WiCell Institute) were cultured as described previously62,63. Cells were maintained with E8 medium on plates coated with Matrigel and split with 0.5 mM EDTA in DPBS. ATCC performs quality testing to ensure authentication of the HEK293T cell line using short tandem repeat analysis. H9 ES cells (from WiCell) are authenticated by WiCell using G-band karyotyping and short tandem repeat analysis. Genetically edited H9 human ES cells were confirmed by karyotyping. HEK293, SUM159T and H9 cell lines were tested for mycoplasma on a monthly basis using Mycoplasma Plus PCR assay kit (Agilent 302107). Use of H9 cells for this study was approved by the Embryonic Stem Cell Research Oversight Committee (approval number 00051).
Human ES cells with the AAVS1-TRE3G-NGN2 driver64 were differentiated into iNeurons as described previously65. Briefly, stem cells were plated at 2 × 105 cells ml−1 (differentiation day 0) in ND1 medium (DMEM with F-12, N-2, human 10 ng ml−1 BDNF, 10 ng ml−1 human NT3, non-essential amino acids, 0.2 μg ml−1 human laminin) supplemented with 2 mg ml−1 doxycycline and 10 μM Y27632 (ROCK inhibitor). The next day, the medium was exchanged with ND1 without Y27632. The following day, the medium was replaced with ND2 (neurobasal medium, B27, GlutaMAX, 10 ng ml−1 BDNF, 10 ng ml−1 NT3) supplemented with 2 μg ml−1 doxycycline. Until the experimental day (day 19–21), 50% of the medium was replaced with fresh ND2 every other day. Cells were replated at 4 × 105 cells per well on day 4–6. From day 10, doxycycline was removed from the ND2.
Lentiviral vectors were packed in HEK293T cells (ATCC number CRL-3216; RRID:CVCL_0045) as described previously62,66,67. Cells were co-transfected at 60% confluency with pPAX2, pMD2 and the target vector in a 4:2:1 ratio using polyethylenimine. The medium was changed to ND2 the next day and collected 2 days after transfection. ND2 medium containing lentivirus was filtered (0.22 μm) and used for transduction of iNeurons at differentiation day 11–12.
CRISPR–Cas9 gene editing
Human ES cells (H9 AAVS1-TRE3G-NGN2 3×Flag–EEA1; RRID:CVCL_D1KV) were gene-edited using CRISPR–Cas9 (ref. 68). Cells were electroporated with a mixture of 0.6 μg guide RNA and 3 μg Cas9-NLS (QB3 MacroLab, University of California, Berkeley) using a Neon transfection system as previously described69 according to the specific protocol at ref. 70. To generate human ES cells homozygous for the TMEM230X121W variant, a single-stranded DNA oligonucleotide was included in the electroporation (5′-CTACCGTGGTTACTCCTATGATGACATTCCAGACTTTGATGACTGGCACCCACCCCATAGCTGAGGAGGAGTCACAGTGGAACTGTCCCAGCTTTAAGATATCTAGCAGAAACTATAGCTG-3′). The cells were recovered for 24–48 h in a low-O2 incubator and sorted into single cells with a Sony Biotechnology (SH800S) cell sorter (RRID:SCR_018066). Gene editing of individual clones was verified by sequencing with the Illumina MiSeq system (RRID:SCR_016379) and validated by immunoblotting and/or MS. Guide RNAs were generated using the GeneArt Precision gRNA synthesis kit (Thermo Fisher Scientific) for the sequences: TMEM230−/− 5′-CCTGAAGGTCAATGTAGCCATCGT-3′, TMEM230X121W 5′-CTCCTCCTCAGCTATGGGGT-3′, TMEM9−/− 5′-TATCTTTGGTGGCTGTGGTC-3′, TMEM9B−/− 5′-TCTACATCAGGCCCCCGCAC-3′. ES cells reported here will be made available upon request, but require a Material Transfer Agreement from WiCell.
Spinning-disc confocal microscopy
For immunofluorescence staining, SUM159PT cells were fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. Cells were blocked with 3% BSA in PBS with 0.1% Triton X-100 for 1 h at room temperature. Cells were incubated with primary antibodies (1:200 dilution) in 3% BSA in PBS with 0.1% Triton X-100 for 3 h at 4 °C. After washes, cells were incubated with Alexa Fluor secondary antibodies (1:400) for 1 h at 4 °C, and nuclei were stained with Hoechst 33342 (1:10,000) for 5 min. Cells were washed and maintained in PBS until microscopy analysis. Immunostaining of iNeurons was performed according to the protocol at ref. 71.
Cells were imaged using a Yokogawa CSU-X1 spinning-disc confocal on a Nikon Eclipse Ti-E motorized microscope and a Plan Apochromat 100× 1.45 N.A oil-objective lens. Live-cell imaging was performed with a Tokai Hit stage top incubator at 37 °C, 5% CO2 and 95% humidity. Images were acquired with a Hamamatsu ORCA-Fusion BT CMOS camera (6.5 μm2 photodiode, 16-bit) and NIS-Elements image acquisition software (RRID:SCR_002776). All samples were measured under the same exposure time and laser power. Co-localization analysis was performed with the JACoP plugin (RRID:SCR_025164) for ImageJ/FiJi (RRID:SCR_002285)72 using maximum-intensity projection images and maximum entropy threshold. Linear mixed-effect model statistics were applied as implemented in the lme4 R package with a nested design to account for images acquired from the same culture well and same biological replicate. The number of fields of view for each of the three independent biological replicates is indicated in the figures (Fig. 4e,f).
Endosomal scoring method
The scoring method was performed to define the endosomal proteome and assign an unbiased score to each protein reflecting the probability of being located in endosomes based on experimental data. The literature was surveyed for studies capturing the endosomal proteome in mammalian organisms, which resulted in 16 datasets18,19,73,74,75,76,77,78,79,80,81,82 (Supplementary Table 1). Incomplete datasets or with ambiguous organelle purifications (for example, ‘vesicles’ or mixed organelles) were excluded. Outdated Uniprot IDs and obsolete gene names were updated (Uniprot 2022-02). Ensembl and the BiomaRt R package (RRID:SCR_019214) were used to retrieve and match rodent genes to their human orthologues, including all human genes when multiple genes matched. Subsequent analyses were based on the protein identification across datasets as a metric for the scoring method (Supplementary Table 1). To evaluate the performance of scoring metrics and datasets, a reference list of 292 well-known endosomal proteins was manually curated from published literature1,3,22,56,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116 (Extended Data Fig. 1a). Dataset overview was visualized by multiple correspondence analysis using the FactoMineR R package (RRID:SCR_014602; Extended Data Fig. 1b). Protein annotation to various organellar locations was based on a previous study62 (Extended Data Fig. 1c). Another metric of the endosomal scoring was the protein abundance in Endo-IP obtained from the label-free proteomic analysis of endosomal pellets as described below (Extended Data Fig. 1d and Supplementary Table 1). The number of interactions with endosomal proteins was obtained from BioPlex 3.0 (RRID:SCR_016144), STRINGDB and CORUM (28.11.2022 Corum 4.1 release)28,117,118 for the reference list of well-known endosomal proteins described above. For STRINGDB, only physical interactions with experimental evidence or databases with high score (combined score >0.7) were included.
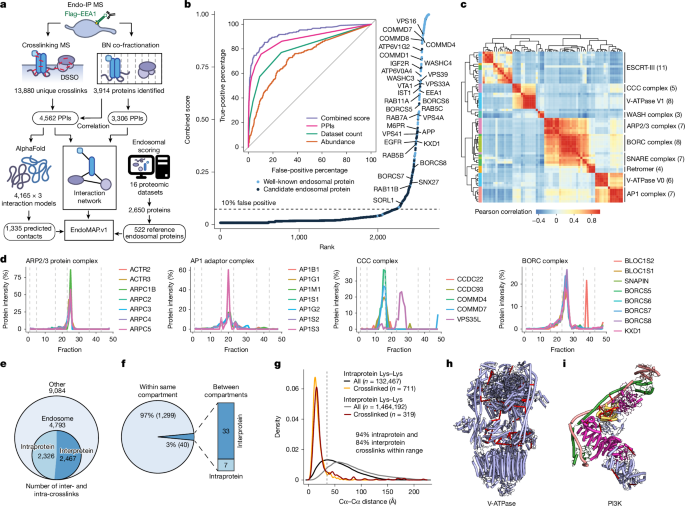
The performance of each metric to classify endosomal proteins (from the reference list described above) was evaluated by receiver operating characteristic curves using the pROC R package with a binomial logistic regression as the predictor (Fig. 1b). The combined endosomal score was obtained by summation of the three individual metrics. Partial area under the curve and the threshold to consider a protein as endosomal was 10% false positives based on the reference list. The scoring method resulted in 407 predicted endosomal proteins (14 proteins present in MitoCarta3.0 (RRID:SCR_018165) were excluded) that were combined with the reference list of well-known endosomal protein for a total of 522 proteins (Supplementary Table 1). These proteins were characterized using BioPlex 3.0 (ref. 28), OpenCell (RRID:SCR_021870)119, and publications as retrieved from Uniprot (Extended Data Fig. 1e–g). Endosomal annotation for all subsequent analyses was based on this list.
EEA1+ endosome purification through Endo-IP affinity capture
Endo-IPs with HEK293EL cells were performed as described previously120. HEK293EL cells expressing Flag–EEA1 (ref. 19) were collected from five 24.5-cm square culture dishes per replicate for co-fractionation experiments (n = 3) and 60 square plates per replicate (divided into two batches) for crosslinking experiments (n = 2). Endo-IPs in iNeurons were performed as described previously44,121. Three 15-cm culture dishes per replicate were used for experiments in iNeurons (n = 3). Cells were pelleted at 1,000g for 2 min at 4 °C and washed once with KPBS buffer (100 mM potassium phosphate, 25 mM KCl and protease inhibitor cocktail, pH 7.2). Cell pellets were resuspended in KPBS and lysed in a Dounce homogenizer with 25 strokes. Samples were centrifuged twice at 1,000g for 5 min at 4 °C, and PNS protein concentration was quantified and normalized by Bradford assay. Samples were incubated for 50 min at 4 °C with 70 μl anti-Flag Sigma magnetic beads for iNeurons experiments, 1.6 ml Sigma anti-Flag Sigma magnetic beads for co-fractionation experiments and 20 ml of anti-Flag Pierce magnetic beads per batch for crosslinking experiments. The beads were washed four times using a magnetic stand with KPBS. For quantitative proteomics, endosomes were eluted with 120 µl 0.5% NP40 (IGEPAL) in KBPS for 30 min at 4 °C and stored at −80 °C until MS sample preparation. For co-fractionation and crosslinking experiments, endosomes were eluted twice with 0.8 mM 3×Flag peptide in KPBS for 45 min at 4 °C (Extended Data Fig. 1h). Peptide-eluted samples were centrifuged for 20 min at 10,000g in Posi-Click tubes (Denville, c2170). Endosomal pellets were washed twice with KPBS to remove excess 3×Flag peptide and immediately processed. An additional wash was performed for the second replicate of the crosslinking experiment, which helped increase the coverage in the MS analysis.
Protein co-IP
A protocol for this analysis is available at ref. 122. Proteins from a 10-cm culture dish of HEK293 cells or a 15-cm dish of iNeurons per replicate (n = 2 or 4) were extracted for 1 h at 4 °C with 0.5% DDM in 25 mM HEPES pH 7.4, 150 mM NaCl and protease inhibitor cocktail123. Samples were centrifuged twice at 20,000g for 20 min, and the supernatant was incubated with 15 μl anti-HA magnetic beads (Pierce) or 25 µl anti-Flag magnetic beads (Sigma) depending on the protein tag for 2 h at 4 °C. For IP using endogenous antibodies, the supernatant was incubated overnight with 5 μg of antibody before the incubation with 15 μl magnetic A/G beads. The beads were separated with a magnetic stand and washed four times with washing buffer (0.1% DDM, 25 mM HEPES, 150 mM NaCl, pH 7.4). Proteins bound to the beads were eluted with 30 μl 1.5× Laemmli buffer for immunoblotting or 30 μl 1.5× S-Trap lysis buffer (7.5% SDS, 150 mM TEAB pH 8.5) for MS analysis and heated at 80 °C for 5 min.
SDS–PAGE immunoblotting
Samples mixed with Laemmli buffer were incubated at 80 °C for 5 min and loaded in a Criterion TGX stain-free precast gel for subsequent immunoblotting. After electrophoresis, gels were scanned using a Bio-Rad ChemiDoc imager (Bio-Rad) and electro-transferred onto a PVDF membrane overnight at 10 V. Membranes were blocked with 5% non-fat milk, and incubated with primary antibody for 2 h at 4 °C and subsequently with HRP-conjugated secondary antibodies for 1 h at 4 °C. After washing, blot images were acquired in a Bio-Rad ChemiDoc imager using SuperSignal West Pico PLUS Chemiluminescence substrate (Thermo Fisher, catalogue number 34580). Images were processed with Bio-Rad Image Lab software (version 6.1.0; RRID:SCR_014210). Differences in loading were normalized using the stain-free quantification of total protein amount. Protocols for this procedure are available at ref. 124. Full versions of all gels and blots are available in Supplementary Fig. 1.
BN electrophoresis co-fractionation and in-gel digestion
A detailed protocol for this procedure is available at ref. 125. Protein complexes from three independent biological Endo-IP replicates were fractionated and processed as previously described126. Freshly prepared purified endosomal pellets were resuspended in 40 μl KPBS with 0.5% DDM, and proteins were extracted for 45 min at 4 °C in rotation. Protein extracts were clarified by centrifugation at 20,000g and mixed with 10 μl BN loading buffer, 1 μl Coomassie G-250 mix and 0.5 μl native molecular weight marker. Samples were run on a 4–16% NativePAGE gel at 150 V for 1.5 h and at 250 V for 20 min at 4 °C. Gels were fixed in 50% ethanol and 3% phosphoric acid, followed by staining with Coomassie. Each sample was cut into 48 1-mm slices and transferred to a 96-well filter plate for in-gel digestion123. Briefly, proteins were reduced with 100 µl 5 mM TCEP in 50 mM ammonium bicarbonate for 30 min at 37 °C. Proteins were alkylated with 20 mM chloroacetamide in 50 mM ammonium bicarbonate for 15 min at room temperature. Fractions were destained, dried and digested with 0.2 μg Lys-C for 4 h at 37 °C followed by overnight incubation with 0.2 μg of trypsin. Peptides were extracted, dried in a SpeedVac and reconstituted in 5% acetronitrile (ACN), 5% formic acid for data-independent acquisition (DIA) liquid chromatography (LC)–MS/MS analysis.
Crosslinking and strong cation exchange fractionation
A detailed protocol for both crosslinking procedures is available at ref. 127. Freshly prepared purified endosomal pellets from two independent biological replicates were resuspended in 300 μl KPBS and immediately crosslinked by incubating with 1 mM DSSO (disuccinimidyl sulfoxide, with the full chemical name bis(2,5-dioxopyrrolidin-1-yl) 3,3′-sulfinyldipropionate, bis-(propionic acid NHS ester)-sulfoxide, Thermo Fisher Scientific) at room temperature for 40 min (ref. 6). The reaction was quenched with 50 mM Tris buffer pH 7.5 at room temperature for 30 min. Crosslinked samples were denatured in 8 M urea, reduced with 5 mM dithiothreitol for 30 min at 37 °C, and alkylated with 40 mM chloroacetamide for 30 min at room temperature. Crosslinked proteins were digested with Lys-C (1:75) at 37 °C overnight. Sample urea concentration was diluted to 2 M with 50 mM 3-[4-(2-hydroxyethyl)-1-piperazine]propanesulfonic acid and incubated at 37 °C with trypsin (1:100) for 6 h. Peptides were desalted with a 50 mg C8 Sep-Pak solid-phase extraction column, dried and fractionated by strong cation exchange chromatography. A 70-min linear gradient of mobile phase (0.5 M NaCl in 20% ACN, 0.05% formic acid) was used from 0 to 8% in 14 min, to 20% at 28 min, to 40% at 48 min and to 90% at 68 min at a column flow rate of 0.18 ml min−1 in a PolyLC PolySulfoethyl A column (3 μm particle size, 2.1 mm inner diameter and 100 mm length). Fractions were collected every 30 s starting at 35 min for 10 min, and then every minute. Fractions were dried in a SpeedVac and desalted using a C8 StageTip. Around 30 fractions for each sample were reconstituted in 5% ACN, 5% formic acid and analysed by LC–MS/MS.
An additional independent biological replicate of freshly prepared purified endosomal pellet was resuspended in 300 μl KPBS and immediately crosslinked by incubating with a combination of 8 mM DHSO and 16 mM DMTMM at 37 °C for 90 min (ref. 32). Crosslinked samples were denatured in 5% SDS and briefly sonicated, reduced with 5 mM dithiothreitol for 5 min at 55 °C, and alkylated with 20 mM MMTS. Crosslinked proteins were precipitated and subjected to the S-Trap mini-spin column digestion protocol as provided by the manufacturer (see below). Peptides were desalted and fractionated by strong cation exchange chromatography as described above. A total of 30 fractions were analysed by LC–MS/MS.
S-Trap sample preparation
Three independent replicates of PNS samples (10 μg or 50 μg of protein depending on the experiment) and Endo-IP samples were mixed with equal volume of water and subjected to sample preparation. The S-Trap micro-spin column digestion protocol (version 4.7) was followed as provided by the manufacturer (Protifi, C02-micro-80)128,129,130. Briefly, each sample was mixed with equal volumes of 2× lysis buffer (10% SDS, 100 mM TEAB buffer pH 8.5). Protein IP samples from iNeurons (n = 2 or 4) were directly collected in 1.5× lysis buffer. Proteins were reduced by incubating at 55 °C for 30 min with 5 mM TCEP and alkylated for 30 min at room temperature with 40 mM chloroacetamide. Samples were acidified with phosphoric acid and mixed with washing buffer (90% methanol, 100 mM TEAB buffer pH 7.55). Samples were transferred to micro-spin columns and washed 4 times with 150 µl washing buffer by centrifugation. Proteins were digested with 0.5 µg Lys-C at 37 °C overnight in a humid chamber, followed by 6 h incubation with 0.5 μg of trypsin. Peptides were collected from the column by three subsequent centrifugation steps (with 50 mM TEAB buffer, 0.2% formic acid and 50% ACN, respectively) and dried in a SpeedVac.
TMT labelling and peptide fractionation
Protocols for labelling of peptides are available at ref. 131. Peptides were resuspended in 50 μl (PNS samples) or 35 μl (Endo-IP samples) 100 mM TEAB buffer pH 8.5. PNS and Endo-IP peptides were labelled by adding 11 μl or 7 μl ACN, and incubating for 1 h at room temperature with 10 μl or 8 μl of TMTpro reagent (20 mg ml−1 stock in ACN), respectively. The reaction was quenched by adding 10 μl 5% hydroxylamine for 15 min.
For PNS samples, equal peptide amounts for each sample were combined, desalted with a 100 mg C18 Sep-Pak solid-phase extraction column and fractionated by basic pH reversed-phase high-performance LC. Chromatography was performed with a 50-min linear gradient from 5% to 35% ACN in 10 mM ammonium bicarbonate pH 8 at a column flow rate of 0.25 ml min−1 using an Agilent 300 Extend C18 column (3.5 μm particle size, 2.1 mm inner diameter and 250 mm length). The initial 96 fractions collected were combined into 24 fractions, as described previously132. One set of 12 non-adjacent fractions were dried in a SpeedVac and desalted using C18 StageTip. Dried peptides were reconstituted in 5% ACN, 5% formic acid and subjected to LC–MS/MS analysis.
For Endo-IP samples, equal peptide amounts for each sample were combined and fractionated using a high-pH reversed-phase peptide fractionation kit (Pierce) following the manufacturer’s protocol. Eluates were combined into six fractions, dried and desalted using C18 StageTip. Dried peptides were reconstituted in 5% ACN, 5% formic acid and subjected to LC–MS/MS analysis.
For protein IP samples, equal peptide amounts for each sample were combined, dried and desalted using C18 StageTip without further fractionation. Dried peptides were reconstituted in 5% ACN, 5% formic acid and subjected to LC–MS/MS analysis.
LC–MS data acquisition
TMT-labelled samples were analysed using a Vanquish Neo UHPLC system coupled to an Orbitrap Eclipse Tribid mass spectrometer (RRID:SCR_020559) with FAIMS Pro (ref. 131). Peptides were separated on a 100-μm microcapillary column packed with 20 cm of Accucore C18 resin (2.6 μm, 150 Å). A 90-min linear gradient from 5% to 20% ACN in 80 min, to 36% at 83 min, and to 98% at 85 min in 0.125% formic acid was used at 0.3 µl min−1. MS1 spectra were acquired on the Orbitrap (resolution 60,000, scan range 350–1,350 m/z, standard automatic gain control (AGC) target, auto maximum injection time). Peptide fragmentation was achieved by high-energy collisional dissociation (HCD) at 36% normalized collision energy. MS2 spectra were acquired on the Orbitrap (resolution 30,000, isolation window 0.6 m/z, TurboTMT set to All TMT Reagents, first mass 120 m/z, 200% normalized AGC, 120 ms maximum injection time). FAIMS Pro was set to −30, −50 and −70 compensation voltage (CV). Unfractionated samples (protein IPs) were injected twice with FAIMS set to −40, −60 and −80 CV for the second run.
BN-PAGE co-fractionation samples were analysed using an EASY-nLC 1200 system coupled to an Orbitrap Exploris 480 mass spectrometer (RRID:SCR_022215). A 15-cm 100-μm capillary column was packed in-house with Accucore 150 C18 resin (2.6 μm, 150 Å). A 90-min linear gradient from 5% to 20% ACN in 80 min, to 25% at 83 min, and to 98% at 85 min in 0.125% formic acid was used at 0.3 µl min−1. The DIA method consisted of MS2 analysis of overlapping isolation windows of 24 m/z stepped through 390–1,014 m/z mass range for the first cycle and 402–1,026 m/z for the second cycle133. DIA scans were performed with 28% normalized HCD collision energy, 30,000 resolution, 145–1,450 m/z scan range, 1,000% normalized AGC and 54 ms maximum injection time. This was followed by a parent MS1 ion scan (resolution 60,000, scan range 350–1,050 m/z, 100% normalized AGC target, auto maximum injection time).
DSSO-crosslinking samples were analysed using an EASY-nLC 1200 system coupled to an Orbitrap Fusion Lumos mass spectrometer with FAIMS Pro (RRID:SCR_020562). A 90-min linear gradient from 5% to 20% ACN in 80 min, to 25% at 83 min, to 40% at 85 min, and to 98% for 2 min in 0.125% formic acid was used at 0.5 µl min−1. An HCD-MS2 strategy was used4, in which the MS1 spectrum was acquired on the Orbitrap (resolution 120,000, scan range 400–1,600 m/z, standard AGC target, auto maximum injection time). Peptides with charge states 4–8 were fragmented by HCD at 21, 27 and 33% normalized collision energy. MS2 was acquired on the Orbitrap (resolution 60,000, isolation window 1.6 m/z, auto scan range, 200% normalized AGC, 120 ms maximum injection time). FAIMS Pro was set to −50, −60 and −75 CV (ref. 134).
DHSO- and DMTMM-crosslinking samples were analysed using a Vanquish Neo UHPLC system coupled to an Orbitrap Ascend MultiOmics Tribid mass spectrometer with FAIMS Pro. A 90-min linear gradient from 5% to 20% ACN in 80 min, to 25% at 83 min, to 40% at 85 min, and to 98% for 2 min in 0.125% formic acid was used at 0.3 µl min−1. MS1 spectrum was acquired on the Orbitrap (resolution 120,000, scan range 350–1,600 m/z, standard AGC target, auto maximum injection time). Peptides with charge states 4–8 were fragmented by HCD at 21, 27 and 33% normalized collision energy. MS2 was acquired on the Orbitrap (resolution 60,000, isolation window 1.4 m/z, auto scan range, 200% normalized AGC, 120 ms maximum injection time). FAIMS Pro was set to −50, −60 and −75 CV.
Proteomics data analysis
TMT-MS data were processed with MSconverter135 and searched using Comet136 against the human canonical proteome (UniProt Swiss-Prot 2021-11), including reverse sequences and common contaminants. Experiments containing variants of TMEM230 were searched against the human canonical proteome (UniProt Swiss-Prot 2024-01) including an additional sequence of TMEM230 with such variants. Peptide mass tolerance was set to 50 ppm and fragment ion tolerance to 0.02 Da. These wide mass tolerance windows were chosen to maximize sensitivity in conjunction with Comet searches and linear discriminant analysis137. TMTpro labels were set as fixed modification on lysines and peptide N terminus (+304.207 Da), carboxyamidomethylation on cysteines (+57.021 Da) as a fixed modification, and oxidation on methionine residues as a variable modification. Linear discriminant analysis was performed138 and peptide-spectrum matches (PSMs) were filtered to 2% FDR139. TMT-reporter ions were quantified by picking the most intense peaks within 0.003 Da around the theoretical m/z, and corrected for isotopic impurity. Only PSMs with at least 200 total signal-to-noise ratio across all TMT channels and 50% precursor isolation purity were used140. Data summarization, normalization and statistics were performed using MSstats141,142. Peptide-level normalization and imputation were enabled, and the protein summarization method was set to ‘LogSum’ for Endo-IP experiments from iNeurons and to ‘msstats’ for all other experiments. The threshold used to consider significantly regulated proteins was 0.05 q-value and 1.5-fold change. For PNS and Endo-IP experiments with iNeurons, three biological replicates per condition were analysed (Supplementary Tables 4 and 5). For protein IP experiments in iNeurons, four biological replicates were analysed per group (Supplementary Table 5), except for one dataset with some groups containing two replicates given the limitation of the maximum number of TMT channels (Supplementary Table 4). Synaptic Gene Ontology enrichment analysis was performed using SynGO143 (https://www.syngoportal.org/#) using all proteins identified in each experiment as background.
DIA-MS data were analysed using DIA-NN (version 1.8) as previously described144,145. Data were converted to mzML using MSconvert135 with the Demultiplex filter set to Overlap Only (10-ppm mass error). A spectral library was generated from the complete human proteome (UniProt 2022-05) with a precursor m/z range of 350–1,050, precursor charge 2–5 and fragment ion m/z range 145–1,450. Carbamidomethylation, oxidation and N-terminal excision were included as modifications. Search was performed with 10-ppm mass accuracy, match-between-runs enabled and robust LC (high precision) quantification strategy. For Endo-IP protocol optimization samples (Extended Data Fig. 1i–l and Supplementary Table 1), downstream analysis was performed using MS-DAP146. Only peptides quantified in all three replicates per condition (n = 3) were included. Data were normalized with variance stabilization normalization and mode-between protein methods. The DEqMS algorithm was selected for statistical analysis, using a significance threshold of 0.01 FDR-adjusted P-value threshold and log2[fold change] of 3 (Supplementary Table 1). For BN co-fractionation experiments, protein complex analysis was performed with PCprophet25. Three biological replicates were analysed with default parameters, the provided core complexes were used as database and the BN markers were used for collapsing hypothesis to common complexes. As previously described7, co-elution scores (from rf output table) were assigned to each protein pair of the complex and used for downstream analysis. Only complexes with a minimum peak elution at 67-kDa and a maximum of 25 proteins per complex were considered. In addition, we considered only interactions with a score of at least 0.7 in two replicates to recover only high-confidence candidate interactions (Supplementary Table 2). These parameters were selected on the basis of the optimal recovery of protein interactions reported in BioPlex7 (Extended Data Fig. 2b,c). Elution profiles and Pearson’s correlation heat map of selected protein complexes based on CORUM118 were generated using the mean normalized elution profile across replicates (excluding outliers as the most dissimilar fraction to the median).
DSSO crosslinking MS data were analysed using Thermo Proteome Discoverer (version 2.5.0.400; RRID:SCR_014477) with the XlinkX module147,148. Data were searched against the human canonical proteome (Uniprot Swiss-Prot 2022-05). MS2 acquisition strategy was selected with 10-ppm precursor mass tolerance, 20 ppm FTMS fragment mass tolerance and 0.6 Da ITMS fragment mass tolerance. Carbamidomethylation was included as a fixed modification; oxidation and N-terminal acetylation were included as variable modifications. A maximum of three trypsin miscleavages was allowed, and the minimum peptide length was set to 5. FDR threshold was set to 5% and only crosslinks with XlinkX score >40 were considered for downstream analysis (Supplementary Table 2). Protein domain information of all crosslinked positions was retrieved from UniProt (Fig. 1f) and copy numbers were obtained from ref. 18 (Extended Data Fig. 2f). Yeast two-hybrid data were retrieved from ref. 29 and IP data from BioPlex 3.0 (ref. 28; Extended Data Fig. 2j). The co-fractionation of crosslinked protein pairs in the BN dataset was evaluated using SECAT149. Positive and negative interaction networks from CORUM were used as provided. The target network was generated from all of the crosslinking interactions for proteins identified in both crosslink and BN. The following parameters were used to ensure the generation of scores for all target protein pairs: peak picking was set to none, monomer threshold factor to 1, minimum abundance ratio to 0, maximum shift to 48 and maximum q-value of 1. SECAT P values were used for comparison with crosslink data and previously reported interactions from STRINGDB, CORUM and BioPlex 3.0 as described above (Extended Data Fig. 2l–n).
DHSO and DSSO crosslinking MS data were analysed using Scout (version 1.6.2)31. Data were searched against the human canonical proteome (Uniprot Swiss-Prot 2022-05) with default parameters, including 10-ppm error on the MS1 level and 20-ppm error on the MS2 level. Carbamidomethyl (mass 57.02146) and MMTS (mass 45.987721) were included as fixed modifications for DSSO- and DHSO-crosslinked samples, respectively; oxidation and N-terminal acetylation were included as variable modifications. A maximum of three trypsin miscleavages and two variable modifications was allowed and the minimum peptide length was set to 6. The FDR threshold was set to 1% at all levels without separation of crosslink types. The ‘residue pairs’ table was used for downstream analysis (Supplementary Table 2).
DMTMM crosslinking MS data were analysed using pLink2 (version 2.3.11, RRID:SCR_000084)150. Data were searched against the human canonical proteome (Uniprot Swiss-Prot 2022-05) with 15-ppm precursor mass tolerance and 20-ppm fragment mass tolerance. Methylthio(C) was included as a fixed modification; oxidation and N-terminal acetylation were included as variable modifications. A maximum of three trypsin miscleavages was allowed and the minimum peptide length was set to 6. Filter tolerance was set to 10 ppm and separated FDR threshold to 1% at the PSM level. Filtered crosslinked sites were used for downstream analysis (Supplementary Table 2). DMTMM and DHSO crosslinks were mapped to all possible protein interactions defined by DSSO crosslinks considering that each DMTMM or DHSO crosslink could match multiple interactions owing to shared peptide sequences.
EndoMAP.v1 network analysis
A PPI network was generated from all protein pairs identified by crosslink and BN. The network was initially filtered to remove proteins present in the native molecular weight markers (spiked-in proteins used as reference in BN experiments), EEA1 (overexpressed and used as a handle for the endosome affinity purification), UBC (in most cases corresponds to a protein modification rather than a member of a protein complex) and keratins (common contaminant). Network characterization and analysis was performed using the igraph R package (RRID:SCR_021238; Extended Data Fig. 3a–c). Proteins were assigned to subcellular location according to the following annotations: endosomal proteins from our scoring method described above (Supplementary Table 1), Golgi proteins (as curated in ref. 140), lysosomal proteins (bona fide proteins in Table S3 from ref. 151; bona fide and experimentally determined proteins in Table S2 and Table S12 from ref. 152), mitochondrial proteins (from MitoCarta3.0 (ref. 153)) and nuclear proteins (based on Uniprot, proteins exclusively designated with nuclear-related terms such as ‘Nucleus’ and ‘Chromosome’). These annotations and the circlize R package (RRID:SCR_002141) were used to generate the network chord diagram (Extended Data Fig. 3d).
The network centred around endosomal proteins (or EndoMAP.v1) was generated by filtering dubious interactions (that is, nuclear proteins) and including only endosomal proteins (as defined by our scoring method) and their direct interactors. Up to 8.5% of the endosomal interactions involved nuclear proteins (Extended Data Fig. 3d), which may be considered questionable (therefore, were filtered out) and may indicate false connectivity at the PPI level introduced by sample preparation. Second-order interactors of endosomal proteins were included only when connected to at least one direct interactor by crosslink and/or two direct interactors by BN (Extended Data Fig. 3e). The core component of the network (that is, biggest module) was visualized using Cytoscape v3.10.1 (RRID:SCR_003032), and protein communities were detected by unsupervised edge-betweenness analysis (Fig. 2a). Gene Ontology (GO) enrichment analysis was performed for each community using g:Profiler (RRID:SCR_022865) with the whole proteome as background (Supplementary Table 2, including only significant GO Cellular Component, GO Biological Process and CORUM terms with at least two proteins). Path distance analysis between proteins assigned to complexes was based on CORUM and GO:CC (only terms related to protein complexes; Fig. 3b and Extended Data Fig. 3f). Graph rewiring with the same degree distribution (100 permutations) was used as a randomized control (Fig. 3c). Disease over-representation analysis of the endosomal proteome was performed on endosomal proteins as defined by our scoring method and as annotated in GO (GO:0005768, date December 2024). Enrichment analysis for the gene network (DisGeNET)154 was performed as implemented in the DOSE R package. Enrichment analysis for neurodegenerative disorders included autism spectrum disorders, epilepsy and severe neurodevelopmental disorder, and schizophrenia was based on ref. 155, and was performed using the clusterProfiler R package (RRID:SCR_016884) with brain-expressed genes as background. Path distance analysis between proteins linked to neurodegenerative disorders was based on Diseases 2.0 (ref. 156; 2024-02 update; RRID:SCR_015664), Parkinson’s disease reviewed genes16 and Parkinson’s disease genome-wide association studies157,158 (Extended Data Fig. 3j,k).
AF-M, AlphaLink2 and structural modelling
AF-M was run with ColabFold v1.5.2 (ref. 8; RRID:SCR_025453) on 40-GB A100 NVIDIA GPUs for all protein pairs identified by XL–MS and three-clique combinations within EndoMAP.v1 (with a maximum of 3,600 amino acids in total). AF-M version 3 was used with weights models 1, 2 and 4 with three recycles, templates enabled, one ensemble, no dropout, and no AMBER relaxation. The multiple sequence alignments supplied to AF-M were generated by the MMSeq server (RRID:SCR_022962) with default settings (paired + unpaired sequences). SPOC and contact sites were calculated as described previously30,159. The quality of the predictions was considered acceptable with a SPOC > 0.33 for pairwise predictions and at least two interfaces with interface average models >0.5 for timer predictions. AlphaLink2 (https://github.com/lhatsk/AlphaLink) was performed as described previously21 using intraprotein and interprotein DSSO crosslinks. Three predictions for each protein pair were generated with AlphaLink2 by using different seeds.
All PDB structures containing protein pairs identified by XL–MS were retrieved by querying the PDB API for X-ray and cryogenic electron microscopy structures with overall resolutions <3.5 Å. PDB chains were mapped to their corresponding UniProt identifiers with PDB SIFTS API. Crosslinks were mapped onto the AF-M and PDB structures, and crosslinked residues with a maximum Cα–Cα distance of 35 Å were considered to match the crosslinker constraints. For AlphaLink2, the maximum Cα–Cα distance considered was 30 Å for all crosslinkers, a more stringent threshold as DSSO crosslinks were already used to assist the prediction generation. For AF-M and AlphaLink2 predictions, only crosslinked residues with both pLDDTs >70 were considered for distance analysis. Crosslinks involving HSP90AA1 and HSP90AB1, which present a large number of crosslinks, were excluded from the distance distribution plots in AlphaLink2 predictions (Extended Data Fig. 4m–o) to make the analysis more representative of the entire dataset.
The association of mTORC1–ragulator complex with V-ATPase was modelled using HADDOCK2.4 web server160 (RRID:SCR_019091). The crosslinks identified between ATP6V1C1–LAMPTOR2 and ATP6V1C1–LAMPTOR4 were used as unambiguous restraints with an upper distance limit of 23 Å and centre-of-mass restraints enabled. The complete mTORC1–ragulator complex structure (PDB 7UXH)59 was included with selected subunits of V-ATPase owing to the limitation in the maximum number of atoms (PDB 6WM2 chains I and J from ATP6V1E1, chains L and M from ATP6V1G1, chain O from ATP6V1C1, chains 8 and 9 from ATP6V0C)58. The hypothetical model with the best score compatible with the expected membrane topology was selected (cluster 5; Extended Data Fig. 11b). Structure images were generated with PyMOL 2.6.0 (RRID:SCR_000305). All input, parameter and output files are available via Zenodo at https://doi.org/10.5281/zenodo.14679635.
Software and resources
The following software, packages and resources were additionally used for analysis and visualization: Rstudio (2023.06.0 Build 421 with R 4.2.1, RRID:SCR_001905); R package ggplot2 (3.5.1, RRID:SCR_014601); R package RColorBrewer (1.1.3, SCR_016697); R package ggrepel (0.9.5, RRID:SCR_016223); R package dplyr (1.1.4); R package FactoMineR (2.11, RRID:SCR_014602); R package pheatmap (1.0.12, RRID:SCR_016418); R package factoextra (1.0.7, RRID:SCR_016692); R package pROC (1.18.5); R package reshape2 (1.4.4); R package igraph (2.1.2); R package tidyr (1.3.1, RRID:SCR_017102); R package lme4 (1.1.13.5, RRID:SCR_015654); R package ggsignif (0.6.4, RRID:SCR_023047); R package viridis (0.6.5) (RRID:SCR_016696); Adobe Illustrator (26.5); NIAID NIH BioArt Source.
Statistics and reproducibility
Sample size, number of replicates and statistical tests are indicated in the figure legends and corresponding sections of the Methods. Validation and representative experiments in Fig. 3d,h and Extended Data Figs. 5c and 7g were performed once, those in Extended Data Figs. 5e,h,k,m and 8c were performed twice, and those in Fig. 4g and Extended Data Fig. 7f were performed three times, with similar results in independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.