Cloning, expression and purification of human SLC35B1
Human SLC35B1 was previously cloned into the yeast GAL1 inducible vector pDDGFP2 (ref. 45), resulting in a construct containing a C-terminal tobacco etch virus (TEV)–GFP–His8 tag. SLC35B1 variants were constructed using polymerase chain reaction (PCR)-based mutagenesis and recloned, as previously described46. The resulting vectors were transformed into Saccharomyces cerevisiae strain FGY217 (MATα, ura3-52, lys2Δ201 and pep4Δ)47. Cells harbouring GFP-fused SLC35B1-expressing plasmids were incubated in 12 l of URA medium containing 0.1% (w/v) glucose at 30 °C in 2-l shaking flasks. Protein expression was induced by the addition of galactose to a final concentration of 2% (w/v) when the optical density at 600 nm reached 0.6. After a 22-h incubation at 30 °C, the cells were harvested, resuspended in a buffer (containing 50 mM Tris-HCl (pH 7.6), 1 mM EDTA and 0.6 M sorbitol) and lysed using a high-pressure cell disruption system (Constant Systems). Membranes were isolated by means of ultracentrifugation at 4 °C and 195,000g for 2 h, homogenized in a buffer (containing 20 mM Tris-HCl (pH 7.5), 0.3 M sucrose and 0.1 mM CaCl2), flash-frozen in liquid nitrogen and stored at −80 °C until use.
SLC35B1 and its variants were purified, as previously described46. In brief, isolated membranes from S. cerevisiae cultures containing GFP-fused SLC35B1 were diluted to a total protein concentration of 3.5 mg ml−1 in a buffer containing 1× PBS, 150 mM NaCl, 10% (v/v) glycerol and 1% (w/v) n-dodecyl-β-d-maltoside (DDM). The membranes were solubilized by mild agitation at 4 °C for 1 h, followed by centrifugation at 120,000g at 4 °C for 45 min to remove the non-solubilized material. Imidazole was added to the supernatant to a final concentration of 10 mM, and the mixture was incubated with 10 ml of Ni2+-nitrilotriacetate affinity resin (Ni-NTA; QIAGEN) for 2 h at 4 °C. The resin was washed with 30 column volumes of a buffer containing 1× PBS, 150 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) DDM and 60 mM imidazole. The protein was eluted in 3 column volumes of 1× PBS, 150 mM NaCl, 0.03% (w/v) DDM and 250 mM imidazole and concentrated to 2 mg ml−1. The concentrate was applied to a PD-10 desalting column (Sephadex G-25; GE) pre-equilibrated in a buffer containing 1× PBS and 0.02% (w/v) DDM. The initial 1.6 ml of the flow-through was collected and concentrated using a 50-kDa molecular weight cut-off spin concentrator (Amicon; Merck Millipore). The same procedure was used to purify each mutant that was fused with GFP. Before large-scale culturing, the membranes of each respective mutant were assessed for monodispersity by fluorescence-detection size exclusion chromatography (FSEC) using a Shimadzu high-performance liquid chromatography (HPLC) LC-20AD/RF-20A (excitation at 488 nm and emission at 512 nm) and Enrich SEC 650 10 × 300 column (Bio-Rad) in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.03% (w/v) DDM.
Thermal shift analysis of SLC35B1–GFP fusion
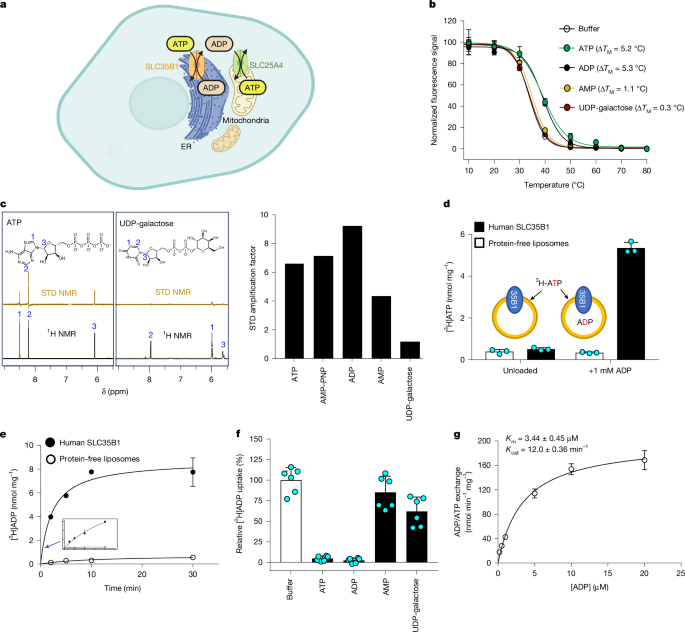
GFP thermal shift experiments were performed, as previously described48,49. In brief, purified SLC35B1–GFP was diluted to 0.6 µM in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl and 1% (w/v) DDM. Substrates of interest were added to the purified SLC35B1–GFP to a final (v/v) concentration of 1 mM, and the resulting mixtures were incubated for 10 min on ice. After incubation, the detergent β-n-octyl-β-d-glucopyranoside was added to a final concentration of 1% (w/v). Subsequently, the samples were transferred to PCR tubes and heated for 10 min at 10, 20, 30, 40, 50, 60, 70 and 80 °C using a Veriti 96-well thermal cycler (Thermo Fisher Scientific). The samples were centrifuged at 5,000g for 45 min at 4 °C to pelletize the larger protein aggregates. The resulting supernatants were transferred to a 96-well black plate (Thermo Fisher Scientific), and GFP fluorescence (excitation at 488 nm and emission at 512 nm) was measured using a Fluoroskan microplate fluorometer (Thermo Fisher Scientific). The apparent Tm for each titration was calculated by plotting the average GFP fluorescence intensity from two technical repeats at each temperature and fitting the curves to a sigmoidal dose–response equation using GraphPad Prism v.8.4. ΔTm was calculated by subtracting the average Tm with nucleotide (calculated from three titrations) from the average Tm without nucleotide (calculated from three titrations).
The relative difference in concentration-dependent ATP-induced thermostabilization was assessed from the final 1 µM to 5 mM, as previously described, but at a single temperature of 37 °C, which was based on wild-type Tm + 4.2 °C, because we previously found a 4–6 °C temperature increase from the Tm was optimal for monitoring ligand binding49. The normalized fluorescence at each concentration of ATP was calculated relative to the GFP fluorescence measured at the starting (lowest) concentration of ATP. Each titration was performed in triplicate.
SLC35B1 proteoliposome transport assays
Purified SLC35B1 was reconstituted into liposomes following the freeze–thaw extrusion method. Lipid extract from bovine brain 7 (Sigma-Aldrich) and cholesteryl hemisuccinate (Sigma-Aldrich) were added at final concentrations of 30 and 5 mg ml−1, respectively, in a buffer containing 10 mM Tris-HCl (pH 7.5) and 2 mM MgSO4. To preload liposomes with nucleotides, 5 µl of a 0.1 M nucleotide stock in 0.5 M Tris (pH 7.5) was added to a 500-µl lipid mix, yielding a final nucleotide concentration of 1 mM. The mixture was flash-frozen and thawed at room temperature before sonication to make unilamellar liposomes. Then 10–20 µl of protein (20 µg total) was added to the liposomes, which were then extruded (LiposoFast; AVESTIN; 400-nm membrane pore size), resulting in large unilamellar proteoliposomes. Liposomes were then diluted in 25 ml of buffer containing 100 mM Tris-HCl (pH 7.5) and 2 mM MgSO4 (transport buffer) and pelleted at 250,000g for 45 min to remove free nucleotides. Finally, the proteoliposomes were resuspended in a transport buffer to a final concentration of approximately 60 mg ml−1. Liposomes without protein (protein-free liposomes) were prepared in the same way but with the addition of the same volume of buffer instead of protein.
To calculate the protein reconstitution efficiency, 120 µl of proteoliposomes and protein-free liposomes at 60 mg ml−1 were solubilized in 1× PBS, 150 mM NaCl (pH 7.5) and 1% (w/v) DDM to a final volume of 300 µl for 1 h at 4 °C. Non-solubilized material was pelleted at 250,000g for 45 min at 4 °C, and the resulting supernatant was injected into an ENrich SEC 650 10 × 300 Column (Bio-Rad) pre-equilibrated with 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.03% (w/v) DDM and ran at 1 ml min−1 using a high-performance liquid chromatography system (Shimadzu) in the same buffer. In addition, 0.6 µg of purified SLC35B1 was injected onto the same column and run as stated previously. This amount of protein should represent 100% theoretical protein reconstitution. Area under the curve (AUC) was obtained from the size exclusion chromatography traces using the AUC function (GraphPad Prism). The AUC values from empty liposomes were subtracted from the AUC proteoliposomes and normalized to the control injection of 100% protein reconstitution. An estimated protein reconstitution of 11% was calculated and used for subsequent calculation of kinetic analysis.
For uptake measurements, 5 µl of proteoliposomes was diluted into 45 µl of transport buffer with either [3H]ATP (0.14 μM) (American Radiolabeled Chemicals and Moravek Biochemicals) or [3H]ADP (0.3 μM) (American Radiolabeled Chemicals) and incubated at 25 °C. Transport was stopped by the addition of 1 ml of transport buffer and by rapid filtration through a 0.22-µm mixed cellulose hydrophilic filter (Millipore). Filters with liposomes were then washed with 6 ml of transport buffer, transferred to scintillation vials and emulsified in 5 ml of Ultima Gold scintillation liquid (PerkinElmer) before scintillation counting (TRI-CARB 4810TR 110 V; PerkinElmer). For the IC50 data acquisition, disintegrations per minute values recorded from protein-free liposomes after 2 min were used for baseline subtraction of the respective tested conditions, and the data were later internally normalized. The IC50 values were obtained by fitting a nonlinear regression of [inhibitor] versus the normalized response with a variable slope using GraphPad Prism v.8.4.
For competitive uptake assays, the uptake of external [3H]ADP (0.14 μM) was monitored in the presence of 1 mM cold nucleotide in a transport buffer after 60 s. For kinetic analysis, the initial velocities were estimated from the initial 30 s of the time-course experiment. A mixture of radiolabelled ADP and ADP at a 1:18 molar ratio was used for the initial points of the curve (0.2–5 µM), and ratios of 1:36 and 1:64 were used for the last points (10 and 20 µM). These different ratios were later corrected when transforming raw radioactive ADP counts to the amount (pmol) of ADP transported. For each concentration of ADP, the disintegrations per minute values from protein-free liposomes were subtracted from their respective proteoliposome values. Final Km and Kcat values were obtained by fitting Michaelis–Menten kinetics using GraphPad Prism v.8.4.
STD NMR measurements
NMR samples were prepared as a mixture of 10 μM purified SLC35B1–GFP into proteoliposomes consisting of total bovine brain lipid extract 7 (Sigma-Aldrich), cholesteryl hemisuccinate (Sigma-Aldrich) and 500 μM of the respective substrate, which were pre-dissolved in a buffer in D2O containing 25 mM potassium phosphate (pH 8.2) and 50 mM NaCl. All NMR experiments were performed at 298 K on a Bruker 500 or 700 MHz spectrometer equipped with cryogenic probes. The NMR spectra were processed using the TopSpin software (Bruker). On- and off-resonance irradiations were applied at chemical shifts of −0.5 and 60 ppm, respectively. Proteins were saturated using a train of Gaussian-shaped 50-ms-long pulses. The total length of the saturation train was set to 2 s. All NMR spectra were acquired with 4,096 scans per dataset. The STD amplification factors were calculated using the following equation to compare the binding intensities22,50.
$${\rm{S}}{\rm{T}}{\rm{D}}\,{\rm{a}}{\rm{m}}{\rm{p}}{\rm{l}}{\rm{i}}{\rm{f}}{\rm{i}}{\rm{c}}{\rm{a}}{\rm{t}}{\rm{i}}{\rm{o}}{\rm{n}}\,{\rm{f}}{\rm{a}}{\rm{c}}{\rm{t}}{\rm{o}}{\rm{r}}={I}_{0}-{I}_{{\rm{s}}{\rm{a}}{\rm{t}}}/{I}_{0}\times {\rm{l}}{\rm{i}}{\rm{g}}{\rm{a}}{\rm{n}}{\rm{d}}\,{\rm{e}}{\rm{x}}{\rm{c}}{\rm{e}}{\rm{s}}{\rm{s}}$$
where I0 are integrated peaks in off-resonance spectra, and I0 − Isat are integrated peaks of the STD spectra.
Pooled CRISPR–Cas9 screen
HCT 116 (Research Resource Identifier: CVCL_0291) cells were transduced in triplicates with lentiviral particles containing a transporter-focused CRISPR–Cas9 library (Addgene, 213695) at a multiplicity of infection of 0.3. Cells were selected with blasticidin for 13 days to remove non-transduced cells and passaged in Roswell Park Memorial Institute 1640 (R8758, Sigma) supplemented with 10% fetal bovine serum (10270-106, lot 42F8381K, Gibco) and penicillin–streptomycin (15140-122, Gibco) for 5 weeks. We performed genomic DNA purification, PCR amplification of the single guide RNA (sgRNA) regions and Illumina sequencing, as previously described51. The sgRNA sequences were quantified using MAGeCK count v.0.5.9.2 (ref. 52). Only sgRNAs targeting SLC transporters and control genes were included in further analysis. Raw count tables were used to determine the significant depletion and enrichment of sgRNAs from the pool using the MAGeCK test v.0.5.9.2 with default parameters. The data analysis was performed on the Galaxy platform53. Raw sequencing data were deposited in the Gene Expression Omnibus (GEO) (GSE277685). The HCT 116 (CCL-247) cell line was purchased from the American Type Culture Collection and authenticated by means of short tandem repeat profiling. PCR testing confirmed the absence of Mycoplasma infection.
Generation of an antibody-based fiducial marker for cryo-EM
All animal experiments conformed to the guidelines of the Guide for the Care and Use of Laboratory Animals of Japan and were approved by the Kyoto University Animal Experimentation Committee. Full-length human SLC35B1 containing residues 1–322 (UniProt accession number P78383) was expressed in the Sf9-baculovirus system and purified. Mouse monoclonal antibodies against SLC35B1 were raised essentially, as previously described54. In brief, a proteoliposome antigen was prepared by reconstituting purified SLC35B1 at a high density into phospholipid vesicles consisting of a 10:1 mixture of chicken egg yolk phosphatidylcholine (egg PC; Avanti Polar Lipids) and adjuvant lipid A (Sigma-Aldrich) to facilitate an immune response. MRL/lpr mice were immunized with proteoliposome antigen using three injections at 2-week intervals. Antibody-producing hybridoma cell lines were generated by using a conventional fusion protocol. Biotinylated proteoliposomes were prepared by reconstituting SLC35B1 with a mixture of egg yolk phosphatidylcholine and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (16:0 biotinyl Cap-PE; Avanti) and used as binding targets for conformation-specific antibody selection. The targets were immobilized on streptavidin-coated microplates (Nunc). Hybridoma clones producing antibodies recognizing conformational epitopes in human SLC35B1 were selected using an ELISA on immobilized biotinylated proteoliposomes (liposome enzyme-linked immunosorbent assay), allowing positive selection of antibodies that recognized the native conformation of SLC35B1. Further screening for reduced antibody binding to SDS-denatured SLC35B1 was performed for negative selection against linear epitope-recognizing antibodies. The stable complex formation between SLC35B1 and each antibody clone was checked using FSEC55. A monoclonal antibody (clone number YN4027) that specifically binds to and stabilizes the conformational epitopes in SLC35B1 was selected. The sequence of Fab YN4027 was determined by means of standard 5′-rapid amplification of cDNA ends using the total RNA isolated from hybridoma cells.
The Fab molecules have a pseudo-symmetrical axis. When the angle between the Fab and target membrane protein is perpendicular, it can be difficult to align the particles correctly in the detergent. To overcome this problem, we created an asymmetric fiducial marker with a single synthetic polyprotein consisting of the YN4027 variable-light domain, short linker, MBP, another linker and YN4027 variable-heavy domain. The resulting Fv–MBP fusion protein was used as a cryo-EM fiducial marker for SLC35B1.
The sequence of the Fv–MBP fusion protein is as follows. The linkers are underlined, and MBP is italicized.
DIVMTQSPASLTVSLGQSVTISCRASENVEYYGTSLMQWYQQKPGQPPKFLIYGASNIESGVPARFSGSGSGTDFSLNIHPVEEDDIAMYFCQQSRKVPYTFGSGTKLEIKGSGKIEEGKLVIWINGDKGYNGLAEVGKKFEKDTGIKVTVEHPDKLEEKFPQVAATGDGPDIIFWAHDRFGGYAQSGLLAEITPDKAFQDKLYPFTWDAVRYNGKLIAYPIAVEALSLIYNKDLLPNPPKTWEEIPALDKELKAKGKSALMFNLQEPYFTWPLIAADGGYAFKYENGKYDIKDVGVDNAGAKAGLTFLVDLIKNKHMNADTDYSIAEAAFNKGETAMTINGPWAWSNIDTSKVNYGVTVLPTFKGQPSKPFVGVLSAGINAASPNKELAKEFLENYLLTDEGLEAVNKDKPLGAVALKSYEEELVKDPRIAATMENAQKGEIMPNIPQMSAFWYAVRTAVINAASGRQTVDEALKDAQTNALGSGEVQLQESGPGLVKPSQSLSLTCSVTGYSITSDYYWNWIRQFPGNKLEWMAYIRYDGTSDYNPSLKNRISITRDTSKNQFFLKLNSVATEDTATYYCARAYYYDGINFDYWGQGTTLTVSSENLYFQ.
The Fv–MBP fusion protein was produced by secretion from the gram-positive bacterium Brevibacillus choshinensis. The DNA sequence of Fv–MBP was inserted downstream of and in frame with the secretion signal sequence of the plasmid pNY326 (Takara Bio/Clontech). To facilitate purification of the secreted proteins, the TEV protease cleavage site sequence, His6 tag and HA tag were added at the C-terminal. B. choshinensis cells harbouring the Fv–MBP expression plasmid were grown at 30 °C with shaking at 200 rpm in 2SY medium (40 g l−1 of soytone, 5 g l−1 of yeast extract, 20 g l−1 of glucose and 0.15 g l−1 of CaCl2) supplemented with 50 mg l−1 of neomycin for 65–70 h. The recovered culture supernatant was adjusted to a final ammonium sulfate concentration of 60% saturation. The resulting precipitate was pelleted, dissolved in Tris-buffered saline (TBS) buffer (10 mM Tris-HCl (pH 7.5) and 150 mM NaCl) and dialysed overnight against the same buffer. The dialysed sample was purified using Ni-NTA resin, mixed with TEV-His6 and dialysed overnight again against the TBS buffer. The cleaved His6-HA tag and TEV-His6 were removed using a HisTrap column. The flow-through fractions were further purified using a HiLoad 16/600 Superdex 75 pg column (Cytiva) equilibrated with the TBS buffer. The peak fractions were pooled, concentrated, flash-frozen in liquid nitrogen and stored at −80 °C.
Cryo-EM sample preparation and data acquisition
For cryo-EM sample preparation, the purified SLC35B1–GFP fusion was incubated at 4 °C overnight with equimolar TEV protease during dialysis in a 3-l buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl and 0.006% (w/v) glyco-diosgenin (GDN). The dialysed mixture was applied to a 5-ml HisTrap HP column pre-equilibrated with 20 mM HEPES (pH 7.5), 150 mM NaCl, 15 mM imidazole and 0.006% GDN, and the flow-through was collected, concentrated to around 2 mg ml−1, flash-frozen and stored at −80 °C.
The purified SLC35B1 protein was incubated on ice with Fv–MBP antibody at a molar ratio of 1:1.2 for 30 min. The complex was isolated by size exclusion chromatography in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl and 0.006% (w/v) GDN detergent. Peak fractions corresponding to the SLC35B1–Fv–MBP fusion complex were concentrated to 5.7 mg ml−1 (wild type), 7.1 mg ml−1 (E33A) and 8.0 mg ml−1 (Q113F) using a 100-kDA cut-off centricon. For data collection with nucleotides, either 5 mM AMP–PNP or 5 mM ADP was added to the protein–Fv–MBP complex at 4 °C before blotting. The concentrated protein sample (3 µl) was applied to either QUANTIFOIL Cu R2/1 (wild type and Q113F) or QUANTIFOIL Cu R1.2/1.3 (E33A) grids and blotted for the optimal time for each construct (3 s for wild type and 1.5 s for E33A and Q113F) at 4 °C under 100% humidity and plunge frozen in liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific).
Cryo-EM datasets were collected using a Titan Krios G3i microscope equipped with a Gatan BioQuantum K3 detector in the super-resolution hard-binned mode. The videos were collected at ×130,000 magnification with aberration-free image shift and fringe-free imaging using EPU (Thermo Fisher Scientific). The other data collection parameters are summarized in Extended Data Table 1.
Cryo-EM data processing
Image processing for all datasets was performed using the CryoSPARC software56. The video frames were aligned using the Patch Motion correction function, and the contrast transfer function was estimated using the Patch CTF algorithm. For apo data, 31,550 videos were recorded. After the CTF estimation, micrographs with an estimated resolution worse than 5.5 Å were rejected. 2D templates were generated from a set of 1,000 micrographs by means of blob picking and 2D classification. Around 9.9 million particles were extracted after template-based picking from the entire dataset. These particles were then subjected to several rounds of 2D classification. Around 1.1 million particles belonging to good 2D classes were selected and subjected to multimodel ab initio reconstruction. A good class containing 443,730 particles was selected and subjected to another round of hetero-refinement and multimodel ab initio reconstruction to remove the particles corresponding to the junk classes. Finally, 180,530 particles were selected for the final round of non-uniform refinement, resulting in a 3D reconstruction with a gold-standard Fourier shell correlation (FSC) resolution of 3.7 Å. To improve the alignments, the flexible MBP domain was masked out and local refinements were performed, which gradually improved the gold-standard FSC resolution to 3.37 Å.
For the SLC35B1 AMP–PNP-bound structure, 16,532 of 17,914 micrographs had an estimated CTF resolution better than 6 Å and were selected for further image processing. Around 8.7 million particles were extracted after template-based picking and subjected to several rounds of 2D classification. Around 420,000 particles were selected, and the initial 3D volumes were obtained using multi-class ab initio reconstruction. We further cleaned up 218,098 particles corresponding to a good 3D reconstruction using several rounds of hetero-refinement. Finally, 114,510 particles were selected and refined to a high resolution using non-uniform refinement. To improve the map features for the transporter region, the volume corresponding to Fv–MBP was masked out and local refinements were performed. The final reconstruction had an overall resolution of 3.44 Å on the basis of the gold-standard FSC at 0.143.
For the SLC35B1 ADP-bound structure, 25,806 of 28,798 micrographs had an estimated CTF resolution better than 6 Å and were selected for further image processing. Around 4.5 million particles were extracted after template-based picking and subjected to several rounds of 2D classification. We selected 1,082,175 particles and obtained the initial 3D volumes using multi-class ab initio reconstruction. We further cleaned up 665,896 particles corresponding to a good 3D reconstruction using several rounds of hetero-refinement. Finally, 323,707 particles were selected and refined to a high resolution using non-uniform refinement. To further improve the map features, the volume corresponding to MBP was masked before local refinement. The final reconstruction had an overall resolution of 2.85 Å on the basis of the gold-standard FSC at 0.143.
For the SLC35B1(Q113F) AMP–PNP-bound structure, 33,714 of 34,801 micrographs had an estimated CTF resolution better than 6 Å and were selected for further image processing. Around 7.7 million particles were extracted after template-based picking and subjected to several rounds of 2D classification. Around 1.2 million particles were selected, and initial 3D volumes were obtained using multi-class ab initio reconstruction. We further cleaned up 590,082 particles corresponding to a good 3D reconstruction using several rounds of hetero-refinement. Finally, 332,121 particles were selected and refined to a high resolution using non-uniform refinement. To further improve the map features, reference-based motion correction was performed, and the volume corresponding to MBP was masked out before local refinements. The final reconstruction contained 329,709 particles and had an overall resolution of 3.0 Å on the basis of the gold-standard FSC at 0.143.
For the SLC35B1(E33A) AMP–PNP-bound structure, 33,579 of 34,144 micrographs had an estimated CTF resolution better than 6 Å and were selected for further image processing. Around seven million particles were extracted after template-based picking and subjected to several rounds of 2D classification. We selected 658,505 particles and obtained the initial 3D volume using multi-class ab initio reconstruction. We selected 313,397 particles and refined them to a high resolution using non-uniform refinement. To further improve the map features, reference-based motion correction was performed, and the volume corresponding to MBP was masked out before local refinements. A round of hetero-refinement was performed to further clean up the data, and a high-resolution reconstruction containing 223,502 particles was obtained after another round of local refinement. The final maps had an overall resolution of 3.0 Å on the basis of the gold-standard FSC at 0.143. The dataset for the ADP-bound SLC35B1(E33A) structure was also processed using CryoSPARC, as described for the other variants (Supplementary Fig. 6). A 3.16-Å resolution map was initially obtained after the local refinement from 306,109 particles with the transporter protein in an outward open conformation. A 3D variability analysis indicated the presence of several conformations; as such, 3D classification without alignment was performed using some of the frames from 3DVA as input volumes. For E33A, ADP-bound states for both the cytoplasmic-facing and luminal-facing conformations were obtained. MBP was masked, and local refinements were performed. The final reconstructions had overall resolutions of 3.15 and 3.12 Å.
Model building
The predicted AlphaFold 2 (ref. 44) model of human SLC35B1 was in the outward open conformation and showed poor side-chain fitting in the cryo-EM maps. To determine the protein conformational state, de novo model building into the apo SLC35B1 maps was instead performed using ModelAngelo software57. The output model was examined and manually adjusted using Coot58. The structure of the Fv fragment was also built de novo using ModelAngelo57 and manually examined and adjusted using Coot58. Because the density corresponding to MBP was masked during the 3D reconstruction, it could not be confidently built into the final maps. The final model containing the SLC35B1 transporter-Fv fragment was refined using real-space refinement in Phenix59.
For model building of the AMP–PNP-bound structure, the model of the apo SLC35B1 structure was fitted to the map density using the Fit in Map utility of ChimeraX60. AMP–PNP was modelled into the extra non-proteinaceous density present at the binding site using Coot58, and the model was refined in Phenix59 using real-space refinement. As the volume corresponding to Fv–MBP was masked during the 3D reconstruction, only the transporter was built into the AMP–PNP complex structure maps. Model building for the ADP-bound SLC35B1 and AMP–PNP-bound SLC35B1(Q113F) variants was also performed using the apo structure as a starting model, followed by manual adjustment in Coot58. Because MBP was masked during refinement, only Fv and the transporter domains were built. The models were refined in Phenix59 using real-space refinement.
For model building of the luminal-facing AMP–PNP- and ADP-bound SLC35B1(E33A) reconstructions, a model obtained from AlphaFold 2 (ref. 44) was fitted to the maps using ChimeraX60. The models were manually examined and adjusted using Coot58 and refined using Phenix59 real-space refinement. For the E33A-ADP cytoplasmic-facing reconstruction, the model of the SLC35B1 structure was fitted to the map. The model was manually adjusted using Coot and refined using Phenix real-space refinement. As MBP was masked out during data processing, only the Fv fragment was built and refined in the final maps of all the structures of the SLC35B1(E33A). Illustrations of the structure and cryo-EM maps were carried out in PyMOL61 and ChimeraX60.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.