Cell culture and treatments
All cell lines were grown at 37 °C under standard cell culture conditions (humidified atmosphere, 5% CO2) in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% FBS (Corning) and 1% penicillin–streptomycin antibiotics. Stable endogenously tagged U-2 OS PCNA–mEmerald 53BP1–mScarlet cells were maintained in the presence of 4 µg ml−1 blasticidin (InvivoGen) and 500 µg ml−1 geneticin (Gibco). Stable endogenously tagged U-2 OS 53BP1–mScarlet cells33 were maintained in the presence of 400 µg ml−1 geneticin (Gibco). Stable U-2 OS PCNA–mEmerald 53BP1–mScarlet cells with overexpression of HRAS or cyclin E1 and their empty vector controls were maintained in the presence of 4 µg ml−1 blasticidin (InvivoGen), 500 µg ml−1 geneticin (Gibco) and 100 µg ml−1 hygromycin B (Invitrogen). RPE-1 PCNA–mEmerald 53BP1–mScarlet cells were maintained in the presence of 4 µg ml−1 blasticidin (InvivoGen) and 500 µg ml−1 geneticin (Gibco). Inducible cyclin E1 RPE-1 cells expressing endogenously tagged PCNA–mEmerald and 53BP1–mScarlet were maintained in the presence of 4 µg ml−1 blasticidin (InvivoGen), 500 µg ml−1 geneticin (Gibco) and 0.5 µg ml−1 puromycin (Thermo Fisher Scientific). U-2 OS 53BP1-GFP AID-DIvA cells40,59 were maintained in the presence of 1 mM sodium pyruvate (Sigma-Aldrich) and 800 µg ml−1 geneticin (Gibco) and 1 µg ml−1 puromycin (Thermo Fisher Scientific). U-2 OS CDC45-GFP MCM4-Halo cells60 were maintained in the presence of 1 mM sodium pyruvate (Sigma-Aldrich) and the HaloTag ligand JF549 (Promega) was added at a concentration of 200 nM for a duration of 20 min before fixation. U-2 OS and RPE-1 cells were reauthenticated by short tandem repeat (STR) profiling in 2025 with 100% match and no detectable contamination (RRID: CVCL_0042 and CVCL_4388). All cell lines used in this study were grown in sterile conditions and routinely tested for mycoplasma.
The following compounds were used in this manuscript: aphidicolin (Sigma-Aldrich), ATRi (AZ-20, TOCRIS), CDKi RO3306 (Sigma-Aldrich), pevonedistat (MLN4924, Selleckchem), etoposide (Selleckchem), 4-OHT (Sigma-Aldrich), IAA (Sigma-Aldrich) and doxycycline (Sigma-Aldrich). Transfections were performed with Ambion Silencer or Silencer Select siRNAs using Lipofectamine RNAiMAX (Thermo Fisher Scientific) at a final concentration of 25 nM. Negative Silencer Select control NEG1 from Ambion was used as a non-targeting control. The following siRNAs were used (5′–3′): TP53BP1 (s14313; GAAGGACGGAGUACUAAUATT); GMNN (134697; GGAGUCAUUUGAUCUUAUGTT); CDKN1A (s416; GCACCCUAGUUCUACCUCATT); TP53 (s606; GAAAUUUGCGUGUGGAGUATT); and AMBRA1 (s31112; GCUCAACAAUAACAUUGAATT).
Cloning
Cloning was done using chemically competent bacteria generated in-house, derived from Library Efficiency DH5a competent cells (Thermo Fisher Scientific). Primer sequences are provided separately (Supplementary Table 5). Correct cloning and integration into target vectors was confirmed by Sanger sequencing (Microsynth).
Cloning of components for endogenous PCNA tagging
The mEmerald-P2A-BlasticidinR pUC18 template was cloned by a three-piece Gibson assembly. The pUC18 vector was linearized by PCR using primers pUC18_lin_fwd and pUC18_lin_rev. The mEmerald gene was amplified by two rounds of PCR from the mEmerald-C1 plasmid template (Addgene, 53975) using first primers mEm_GA_fwd and mEm_P2A_rev and the product amplified with GA_pUC18_fwd and mEm_P2A_rev. The blasticidin gene was amplified with primers BlastR_P2A_fwd and BlastR_pUC18_rev. The individual pieces were purified by gel extraction and assembled with Gibson assembly followed by transformation of the product, isolation of plasmids and verification by sequencing.
The sgRNA duplex to target Cas9 endonuclease in pX459 (Addgene, 48139) to the endogenous PCNA C-terminal near the stop codon was generated as described61. In brief, primers PCNA_sgRNA_top and PCNA_sgRNA_bot were phosphorylated with T4 phosphonucleotide kinase (NEB) at 37 °C for 30 min followed by a temperature gradient from 95 °C to 25 °C decreasing 5 °C min−1. The product was diluted 100-fold in water and assembled into the vector by Golden Gate assembly using BbsI (NEB) and T4 DNA ligase (NEB) through 12 cycles between 5 min at 37 °C and 5 min at 16 °C followed by transformation, plasmid isolation and sequencing verification of correct insertion.
The repair template for tagging endogenous PCNA by homology-directed repair was amplified by PCR from the mEmerald-P2A-BlasticidinR pUC18 plasmid generated as described above using Q5 (NEB) polymerase amplification with primers PCNA_HDR_mEm_fwd and PCNA_mEm_blst_rev followed by PCR purification with a QIAquick PCR purification column according to the manufacturer’s instructions.
Endogenous 53BP1–mScarlet and PCNA–mEmerald tagging in U-2 OS and RPE-1 cells
U-2 OS cells with endogenously tagged 53BP1–mScarlet were described previously33. RPE-1 cells with endogenously tagged 53BP1–mScarlet were generated in the same manner. These cell lines were then used as the starting point for endogenous tagging of PCNA with the monomeric green fluorescent protein mEmerald. A total of 40,000 cells was seeded into two wells of a six-well plate one day before transfection. The PCR amplified PCNA mEmerald-P2A-BlasticidinR HDR template and the pX459 Cas9 plasmid generated as described above were used for transfection. Then, 1 µg of the HDR template and 1 µg of the pX459 plasmid were diluted with 250 µl OptiMEM (Invitrogen) and mixed with 6 µl TransIT-LT1 transfection reagent (Mirus). pX459 without the HDR template was prepared similarly as a negative control. This was allowed to stand 15 min before adding it to the cells. Next, 24 h later, cells were transferred to 15 cm dishes. Then, 48 h after transfection, blasticidin (InvivoGen) was added to a final concentration of 4 µg ml−1 for selection the next 7 days. After selection and death of cells transfected without the HDR template, individual colonies were picked by trypsin detachment in cloning cylinders and transferred to a 96-well plate (Greiner µ-clear) for expansion and validation of the presence of green fluorescence. Clones with fluorescence were further expanded and cells collected for genomic PCR with the primers PCNA_genCterm_fwd and PCNA_genCterm_rev to confirm the insertion of DNA with a size corresponding to the mEmerald-P2A-BlasticidinR module by agarose gel electrophoreses imaged on Infinity ST5 Xpress v16.16d. To visualize endogenous 53BP1 and endogenous PCNA with minimal interference of their cellular functions, monoallelic targeting was considered sufficient. The mEmerald-C1 plasmid was a gift from M. Davidson (Addgene, 53975), pSpCas9(BB)-2A-Puro (PX459) was a gift from F. Zhang (Addgene, 48139)61, pmScarlet-i_C1 was a gift from D. Gadella (Addgene, 85044)62.
Cloning of pBABE_EV and pBABE_HRAS V12
For retroviral transduction, retroviral vectors pBABEneo-HRASV12, a gift from J. Debnath (Addgene, 71304)63 and pBABEneo were used. For them to be compatible with the stable PCNA–mEmerald 53BP1–mScarlet U-2 OS cells, the resistance of the pBABE plasmids was exchanged from neomycin to hygromycin. The pBABE_EV (EV = empty vector) and pBABE_HRAS V12 backbones were amplified by PCR amplification using primers Backbone_fwd and Backbone_rev while adding AscI and MfeI restriction sites. The HygroR insert was also generated by PCR amplification using primers Hygro_fwd and Hygro_rev, adding AscI and MfeI restriction sites. This was followed by incubation with DpnI (NEB) for 2.5 h at 37 °C. PCR-cleanup was done according to the manufacturer’s instructions (QIAquick PCR Purification Kit, Qiagen). Both the backbones and HygroR inserts were incubated with AscI (NEB) for 10 h, followed by heat inactivation of the restriction enzyme at 80 °C for 20 min. This was followed by incubation with MfeI (NEB) for 10 h at 37 °C. Backbones were dephosphorylated using rSAP (NEB) for 1 h followed by gel purification (QIAquick Gel Extraction Kit, Qiagen). The ligation of the backbones and HygroR inserts was done at 16 °C for 16 h. Transformation was performed using chemically competent DH5α generated in house, derived from Library Efficiency DH5α competent cells (Thermo Fisher Scientific). Correct sequences were confirmed by control digestion and sequencing.
Cloning of pBABE_cyclin E1
The pBABEneo-cyclin E1 plasmid for retroviral transduction was a gift from P. Janscak (University of Zurich). To exchange the resistance from neomycin to hygromycin, the pBABE_cyclin E1 plasmid was linearized using primers pBABE_lin_fwd and pBABE_lin_rev. The insert was generated using the primers pBABE_Gibson_fwd and pBABE_Gibson_rev. After gel extraction, Gibson assembly and transformation were performed. Correct sequences were confirmed by control digestion and sequencing.
Cloning of cyclin E1 plasmid for inducible expression
The pLVX-TetONE plasmid was linearized using the primers pLVX-TetONE_lin_rev and pLVX-TetONE_lin_fwd and the insert for cyclin E1 was amplified with primers pLVX-TetONE_to_cyclinE1_GA_amp_fwd and pLVX-TetONE_to_cyclinE1_GA_amp_rev. After gel extraction, Gibson assembly and transformation were performed. Correct insertion was confirmed by control digestion and sequencing.
Generation of oncogene-overexpressing cell lines
For the generation of U-2 OS PCNA 53BP1 cells with overexpression of HRAS V12 or cyclin E1, the U-2 OS PCNA 53BP1 cells were used for retroviral transduction. For this, HEK293T Phoenix retrovirus producer cells for transduction were prepared by plating them 48 h before infection in DMEM (Gibco) containing 10% FBS (Corning) and 1% penicillin–streptomycin antibiotics (Gibco). For transduction, chloroquine was added to each plate of HEK293T Phoenix cells at a final concentration of 20 µM 1 h before transduction. For the formation of the calcium-phosphate-DNA co-precipitate deionized water, 10 µg DNA and CaCl2 (Sigma-Aldrich) at a final concentration of 1.25 M were mixed for a final volume of 500 µl per 10 cm plate. This mix was added dropwise to 500 µl 2× HBS (50 mM HEPES (ChemieBrunschwig), 10 mM KCl (Sigma-Aldrich), 12 mM dextrose (Sigma-Aldrich), 280 mM NaCl (Merck), 1.5 mM Na2HPO4·7H2O (Merck)) solution while vortexing vigorously. After incubating for 5 min, the H2O/DNA/CaCl2/HBS mix was added dropwise to the HEK293T Phoenix cells and then gently distributed. After 6 h of incubation at 37 °C under standard cell culture conditions (humidified atmosphere, 5% CO2), the medium was exchanged to fresh growth medium. Then, 48 h after transduction, the supernatant from the transfected HEK293T Phoenix cells was collected and centrifuged and the supernatant transferred to a new tube. Polybrene (Sigma-Aldrich) was added to the supernatant at a final concentration of 8 µg ml−1. After removal of the medium from the target cells, the viral supernatant was added. After 3 h, the infection was repeated with fresh viral supernatant. After 24 h, the viral supernatant was exchanged to selection medium (DMEM containing 10% FBS (Corning), 1% penicillin–streptomycin antibiotics (Gibco), 4 µg ml−1 blasticidin (InvivoGen), 500 µg ml−1 geneticin (Gibco) and 100 µg ml−1 hygromycin B (Invitrogen)). After selection, the expression levels and functionality were validated by immunofluorescence staining and western blot analysis.
For the generation of RPE-1 cells with inducible overexpression of cyclin E1, RPE-1 cells expressing endogenously tagged PCNA–mEmerald and 53BP1–mScarlet were subjected to lentiviral transduction. For this, a DNA/CaCl2/H2O mix was prepared by mixing 6 µg of LTR plasmid and 6 µg of pCDN-LBH plasmid with 12 µg of oncogene-encoding plasmid with CaCl2 at a final concentration of 250 mM in H2O. To this mixture, 500 µl 2× HBS (50 mM HEPES (ChemieBrunschwig), 10 mM KCl (Sigma-Aldrich), 12 mM dextrose (Sigma-Aldrich), 280 mM NaCl (Merck), 1.5 mM Na2HPO4·7H2O (Merck)) was added dropwise, while vortexing. After incubating the precipitate for 5 min at room temperature, it was added to HEK293T cells and left overnight at 37 °C and 5% CO2. The next day, the medium was replaced with fresh medium and cells were incubated again overnight. The virus was then collected on two consecutive days, the fractions were pooled and filtered through a 0.45 µm filter and stored at 4 °C. The viral supernatant was diluted with medium 1:2 and added together with 8 µg ml−1 polybrene to the target cells. After overnight incubation, the virus-containing cell supernatant was replaced with fresh medium and cells were incubated for 48 h. Then, puromycin was added at a final concentration of 0.5 µg ml−1 for selection. After selection, the expression levels and functionality were validated by immunofluorescence staining and western blot analysis.
Live-cell imaging and treatments
On the day before imaging, cells were seeded in Fluorobrite DMEM (Thermo Fisher Scientific, A1896701) supplemented with 10% FBS (Corning), penicillin and streptomycin (Gibco) and GlutaMAX (Gibco) into 96-well µ-plates (Ibidi, 89626). In the case of RPE-1 PCNA–mEmerald 53BP1–mScarlet cells, before cell seeding, the 96-well imaging plates were coated with collagen diluted in water (1:50) (PureCol-S, 5015-20ML, Advanced Biomatrix) according to the protocol provided by the company. The plate was sealed with a Breathe-Easy sealing membrane (Sigma-Aldrich, Z380059). Live-cell imaging of cells was done using a previously described automated widefield GE InCell Analyzer 2500HS high-content screening microscope33 with environmental control for gas (5% CO2 and 20% O2) and temperature (37 °C) using the GE InCell Analyzer 2500 V7.4 acquisition software. The system contains a seven-colour solid-state illuminator (SSI), a PCO-sCMOS camera system (16 bit, 2,048 × 2,048 pixel, pixel size 6.5 × 6.5 μm, readout speed: 272 MHz), two quad band-pass polychroic mirrors and single band-pass emission filters. For Live+QIBC experiments, the polychroic beam splitter BGOFR_1 (blue (excitation BP 390/22, emission BP 435/48), green (excitation, BP 473/28; emission, BP 511/23), orange (excitation, BP 542/30; emission, BP 597/45), far red (excitation, BP 631/28; emission, BP 684/24)) and a CFI Plan Apo Lambda ×20 Air objective (NA 0.75, WD 1.0 mm) with hardware autofocus were used. Single-plane widefield images were acquired at 100 ms exposure for green and 300 ms exposure for orange; no binning was performed. Nine fields per well were acquired at 30 min intervals for up to 72 h, with typically 2–3 wells per experimental condition and at least 3,000 cells seeded per well.
To irradiate the cells, the plate acquisition was temporarily paused for irradiation at 130 kvp for 33 s per 1 Gy of irradiation in a Faxitron Cabinet X-ray System Model RX-650. Unless indicated otherwise, irradiation was performed with 4 Gy. The addition of drugs was done likewise. At the end of the live imaging, cells were washed once with PBS followed by fixation with 4% formaldehyde (Sigma-Aldrich) in PBS for 18 min, before being washed once more with PBS followed by storage at 4 °C until multiplex staining.
Multiplex staining
Cells fixed after live-cell imaging were processed for iterative indirect immunofluorescence imaging (4i) as described previously35. The cells were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 5 min followed by washing with PBS. Cells were then stained with DAPI (Thermo Fisher Scientific) for 10 min, washed with PBS and images of the fixed cells with endogenous labels and DAPI were acquired. The signals from the fluorescent proteins were removed by denaturation with elution buffer (0.5 M glycine (BioSolve Chemicals), 3 M urea (Eurobio Scientific), 3 M guanidine hydrochloride (Sigma-Aldrich), 50 mM TCEP (Sigma-Aldrich) in water, pH 2.5) for 10 min, 2 times, followed by three washes with double-distilled H2O. Images were acquired to control for the loss of signal. The cells were then blocked and free cysteines were conjugated by incubation with sBS buffer (1% bovine serum albumin (Sigma-Aldrich), 150 mM maleimide (Sigma-Aldrich) in PBS, pH 7.4) for 1 h. After two washes with PBS the cells were incubated with 100 μl cBS (1% BSA (Sigma-Aldrich) in PBS, pH 7.4) with primary antibodies anti-γH2AX (1:1,000, mouse, BioLegend 613402) and anti-pRb (1:500, rabbit, Cell Signaling Technologies, 8516S) for 2 h. After two PBS washes, cells were incubated with 100 µl cBS with secondary antibodies anti-mouse A488 (1:500, Thermo Fisher Scientific, A11029) and anti-rabbit A647 (1:500, Thermo Fisher Scientific, A21245) for 1 h. Finally, cells were stained with DAPI. Elution and acquisition of images after elution was repeated as above, followed by sBS blocking for 1 h and washing. Then cells were stained with anti-p21 (1:500, rabbit, Abcam, ab109520) and anti-p53 (1:500, mouse, Thermo Fisher Scientific, AHO0152) for 2 h. After two PBS washes, cells were incubated with anti-mouse A488 (1:500, Thermo Fisher Scientific, A11029) and anti-rabbit A647 (1:500, Thermo Fisher Scientific, A21245) for 1 h. For every staining or elution round, imaging buffer (700 mM N-acetyl-cysteine (Sigma-Aldrich) in double-distilled H2O, pH 7.4) was added before imaging. Imaging was performed using the automated widefield GE InCell Analyzer 2500HS high-content screening, which was also used for live-cell imaging (seven-colour SSI, PCO-sCMOS camera system (16 bit, 2,048 × 2,048 pixel, pixel size: 6.5 × 6.5 μm, readout speed: 272 MHz), two quad band-pass polychroic mirrors, single band-pass emission filters). The polychroic beam splitter BGOFR_1 (blue (excitation BP 390/22, emission BP 435/48), green (excitation BP 473/28, emission BP 511/23), orange (excitation BP 542/30, emission BP 597/45), far red (excitation BP 631/28, emission BP 684/24)) and a CFI Plan Apo Lambda ×20 Air objective (NA 0.75, WD 1.0 mm) were used, with hardware laser autofocus for acquisition of single plane images. No binning was performed. Nine fields per well were acquired, with typically 2–3 wells per experimental condition and at least 3,000 cells seeded per well. For all imaging and staining rounds the acquisition settings were kept constant.
Analysis of live-cell imaging with 4i acquisitions after fixation
Conversion of images from live-cell acquisition to multi-parametric time-correlated readouts
Images from the live imaging were processed using a time-lapse script (https://github.com/AltmeyerLab/SingleCellTracking_Timelapse), which involves generation of .tif stacks and generation of Olympus ScanR Analysis (v.3.0.1, 3.2 and 3.3.0) compatible images. This was followed by custom conversion in Olympus ScanR Analysis software (v.3.0.1, 3.2 and 3.3.0) to generate the required metadata and the experiment file. Segmentation of cells was performed on a smoothened mask of the PCNA channel. Parameters for total and mean intensities of the two channels, nuclear area (1 pixel ≅ 0.1056 µm2), circularity factor, perimeter, elongation factor, foci counts for PCNA and 53BP1, integrated intensities of foci in each channel, time slices, well number, position, centre x coordinate and centre y coordinate were extracted and tabulated for each cell in every timeframe.
Alignment and analysis of 4i acquisitions
For alignment of 4i imaging data, an alignment script was used (https://github.com/AltmeyerLab/SingleCellTracking_Multiplex-Alignment), which renders images from multiple rounds of staining compatible with downstream analysis using the Olympus ScanR Analysis software (v.3.0.1, 3.2 and 3.3.0). Custom conversion and image analysis was performed with the Olympus ScanR Analysis software (v.3.0.1, 3.2 and 3.3.0). In addition to the parameters outlined above, the mean and total intensities for all stainings as well as γH2AX foci counts and integrated intensities of γH2AX foci were extracted and tabulated for each cell.
Lineage-based tracking of live cells
Tables with data from a field of view from the post-fixation 4i acquisition were appended to data from the live-cell imaging for the same field of view. The frame numbers and x–y coordinates were used as the basis for a previously published MATLAB (MathWorks, MATLAB R2019b, R2020b, R2023a) script to automatically generate live-cell tracks for each cell. The same script along with the ImageJ/Fiji 64-bit (v.1.53f, 1.53t, 1.54f, 1.54m) plugin ‘Manual Tracking’ was used together with the corresponding image stacks to visualize, correct and manually reassign cells for lineage-based analysis. After reassignment, MATLAB scripts (both scripts are available at GitHub; https://github.com/AltmeyerLab/MatlabTracking/; script 1, Cell_tracking.m; and script 2, KeyDataExtractions.m) were used to generate lineage assignments and data from curated and reassigned cells was written into .csv files and loaded into TIBCO Spotfire (v.7.9.1, 10.10.1) for visualization of the lineage trees together with the post-fixation 4i data. For U-2 OS cells, at least 20 individual cell lineages per condition, corresponding to up to 80 granddaughter cells, were tracked. For RPE-1 cells, at least ten individual cell lineages per condition, corresponding to up to 40 granddaughter cells, were tracked. For all extended live-cell imaging experiments with lineage tracking and multiplex staining (Live+QIBC), at least two independent biological replicate experiments were performed. Area measurements are shown in pixel with 1 pixel ≅ 0.1056 µm2. In some instances, although the median and the complete Q1–Q3 range are shown for whole-population box plots associated with Live+QIBC results, not all individual values are displayed to allow for visualization of population effects with consistently scaled axes.
Cell cycle analysis and analysis of sister cell heterogeneity
After validating that PCNA foci were mostly confined to EdU-positive S-phase cells, thresholds were established for S-phase categorization as well as for G1–S and S–G2 transitions based on live-cell tracking of untreated control cells. In brief, unchallenged cells transitioned from G1 to S phase quickly (typically within a 30-min interval), indicated by a marked increase in PCNA foci (≥10 foci). Likewise, unchallenged cells transitioned from S phase to G2 quickly (typically within 1–2 30-min intervals), indicated by a marked decrease in PCNA foci (≤4 foci). Thresholds were therefore defined as follows: 0–4 PCNA foci for G1; ≥10 PCNA foci for ≥2 consecutive timepoints for S phase (G1–S transition); ≤4 PCNA foci for ≥2 consecutive timepoints for G2 phase (S–G2 transition). Periods in between were considered transition periods.
53BP1 foci heterogeneity was evaluated on the basis of the endogenous 53BP1 foci in the F1 generation measured from the live imaging data. For the other markers, heterogeneity was evaluated from multiplexed end-point measurements. Three categories of sister cell heterogeneity were introduced based on the differences in foci counts or nuclear intensity levels; thresholds were dependent on the individual experiment: for Extended Data Fig. 5e, 53BP1 foci heterogeneity was categorized as follows: low (0–1 focus difference), medium (2–4 foci difference), high (>4 foci difference). γH2AX fluorescence intensity was categorized as follows: low (0–100 a.u. difference), medium (100–200 a.u. difference), high (>200 a.u. difference). p53 fluorescence intensity was categorized as follows: low (0–50 a.u. difference), medium (50–100 a.u. difference), high (>100 a.u. difference). p21 fluorescence intensity was categorized as follows: low (0–100 a.u. difference), medium (100–500 a.u. difference), high (>500 a.u. difference). For Extended Data Fig. 8i, 53BP1 foci heterogeneity was categorized as follows: low (0–1 focus difference), medium (2–4 foci difference), high (>4 foci difference). For γH2AX fluorescence intensity, heterogeneity was categorized as follows: low (0–50 a.u. difference), medium (50–100 a.u. difference), high (>100 a.u. difference). For p53 fluorescence intensity, heterogeneity was categorized as follows: low (0–200 a.u. difference), medium (200–500 a.u. difference), high (>500 a.u. difference). For pRb fluorescence intensity, heterogeneity was categorized as follows: low (0–500 a.u. difference), medium (500–1,000 a.u. difference), high (>1,000 a.u. difference). For Supplementary Fig. 8b, 53BP1 foci heterogeneity was categorized as follows: low (0–1 focus difference), medium (2–4 foci difference), high (>4 foci difference).
High-content microscopy and QIBC
Automated multichannel widefield microscopy for high-content imaging and QIBC was performed using the Olympus ScanR High-Content Screening System as described previously33,64. The system is equipped with an inverted motorized Olympus IX83 microscope, a motorized stage, IR-laser hardware autofocus, a fast emission filter wheel with one set of band-pass filters for multi-wavelength acquisition (DAPI (excitation BP 395/25 or BP 390/22, emission BP 435/26), FITC (excitation BP 470/24 or BP 475/28, emission BP 515/30), TRITC (excitation BP 550/15 or BP 555/28, emission BP 595/40), Cy5 (excitation BP 640/30 or BP 635/22, emission BP 705/72)), and a Hamamatsu ORCA-FLASH 4.0 V2 sCMOS camera (12 bit, 2,048 × 2,048 pixel, pixel size 6.5 × 6.5 μm) with a ×40 UPLSAPO (NA 0.9, WD 0.18 mm), a ×20 UPLSAPO (NA 0.75, WD 0.6 mm) and a ×10 UPLSAPO (NA 0.4, WD 3.1 mm) air objective. Images of cell populations were acquired under non-saturating conditions (Olympus ScanR Image Acquisition software (v.3.0.1, 3.2 and 3.3.0)), typically 25 (5 × 5) to 81 (9 × 9) images per well, depending on the objective and cell density, and identical settings were applied to all samples within one experiment. Hardware and software autofocus on the DAPI channel were used. No binning was performed. Images were analysed using the inbuilt Olympus ScanR Analysis software (v.3.0.1, 3.2 and 3.3.0), a dynamic background correction was applied and nucleus segmentation was performed using an integrated intensity-based object detection module based on the DAPI signal. Downstream analyses were focused on properly detected nuclei containing a 2N–4N DNA content as measured by total and mean DAPI intensities, unless increased ploidy in cells with >4N DNA content was also analysed. Fluorescence intensities were quantified and are depicted as arbitrary units. Colour-coded scatterplots of asynchronous cell populations were generated with TIBCO Spotfire (v.7.9.1, 10.10.1). Within one experiment, similar cell numbers were compared for the different conditions. For visualizing discrete data in scatterplots, mild jittering (random displacement of data points along discrete data axes) was applied to demerge overlapping datapoints. Representative scatterplots and quantifications of independent experiments, typically containing several thousand cells each, are shown.
DNA fibre imaging
A Leica THUNDER (Las X 3.7.6.25997) Imager 3D Live Cell system was used for DNA fibre analysis. The system is equipped with a Leica DMi8 inverted widefield microscope, a motorized stage, a Leica LED8 light source (390 nm, 440 nm, 475 nm, 510 nm, 555 nm, 575 nm, 635 nm, 747 nm) and a Leica monochrome fluorescence DFC9000 GTC sCMOS camera (12/16 bit, 2,048 × 2,048 pixel, pixel size: 6.5 × 6.5 μm). Dual-labelled DNA fibres (Alexa Fluor 488 and 555) were imaged with the 475 nm and 555 nm LEDs, the DFT5 Quad filter (excitation filters: 375–407, 462–496, 542–566, 622–654; main beam splitter: 415, 500, 572, 660; emission filters: 420–450, 506–532, 581–607, 666–724), additional emission filters 535/70 and 642/80 from an external clean-up filter wheel, and a Leica HC PL APO CS2 (NA 1.4, WD 0.14 mm) ×63 oil objective.
Immunostaining
For standard immunofluorescence staining, high-content microscopy and QIBC analyses, cells were seeded on sterile 12 mm glass coverslips or 96-well plates and were allowed to proliferate until they reached a cell density of 70–90%. Cells were then fixed in 3% formaldehyde (Sigma-Aldrich) for 15 min at room temperature, washed once in PBS, permeabilized for 5 min at room temperature in 0.2% Triton X-100 (Sigma-Aldrich) in PBS, washed twice in PBS and incubated in blocking solution (filtered DMEM containing 10% FBS (Corning) and 0.02% sodium azide (Merck)) for 15 min at room temperature. When the staining was combined with an EdU Click-iT reaction, the reaction was performed before the incubation with the primary antibody according to manufacturer’s recommendations (Thermo Fisher Scientific). Where indicated, cells were pre-extracted in 0.2% Triton X-100 (Sigma-Aldrich) in PBS for 2 min on ice before formaldehyde fixation. Denaturing, where indicated, was performed in 2.5 M HCl for 10 min. All primary antibodies were diluted in blocking solution and incubated for 2 h at room temperature. Secondary antibodies (Alexa Fluor 488, 555, 568, 647 anti-mouse, anti-rabbit, anti-goat and anti-rat IgG, Thermo Fisher Scientific) were diluted 1:500 in blocking solution and incubated at room temperature for 1 h. Cells were washed once with PBS and incubated for 10 min with DAPI (0.5 mg ml−1) in PBS at room temperature. After three washing steps in PBS, the coverslips were briefly washed with distilled water and mounted onto 6 µl Mowiol-based mounting medium (Mowiol 4.88 (Calbiochem) in glycerol/Tris), whereas the wells of the 96-well plates were kept filled with PBS.
Immunoblotting
Proteins were separated by standard SDS–PAGE and transferred onto PVDF membranes. Membranes were blocked with 5% milk in PBS-T (PBS + 0.1% Tween-20) for 1 h at room temperature and incubated with primary antibodies over night at 4 °C. The membranes were then washed three times with PBS-T and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature, washed again three times with PBS-T and protein signals were detected using ECL Western Blotting Detection Reagent (Thermo Fisher Scientific) and an OPTIMAX X-Ray Film Processor (PROTEC Medizintechnik). To enable detection of multiple target proteins on the same membrane without stripping (typically 2–3 target proteins per membrane with sufficiently distinct molecular weight), membranes were cut horizontally, each piece was probed with specific primary and secondary antibodies, and the membrane was then reassembled for ECL detection. Original western blot scans are provided in Supplementary Fig. 1.
Clonogenic survival assay
Cells transfected with the indicated siRNAs were seeded at single-cell density and exposed to the indicated drugs at the indicated final concentrations. All conditions were performed in triplicates. Cells were then incubated for 10 days and the number of colonies with more than 50 cells was counted after staining with crystal violet (0.5% crystal violet (Sigma-Aldrich) in 20% ethanol). GraphPad Prism (v.9 and 10) was used to display clonogenic survival data.
scRNA-seq and SORT-seq
For SORT-seq, cells were sorted on a FACS Aria III 5L (FACS Diva Software v.8.0.1) system equipped with a 488 nm laser line. For the polyploidy dataset, pevonedistat-treated cells were treated with 175 nM of pevonedistat for 24 h. Before single-cell sorting, all live cells were incubated for 1 h with a final concentration of 5 µg ml−1 Hoechst 33342 (Thermo Fisher Scientific). All reagents used until the moment of sorting also contained 5 µg ml−1 Hoechst 33342 (Thermo Fisher Scientific). For the scSeq dataset, where IR-treated cells were compared to untreated cells, U-2 OS cells were treated with 4 Gy of IR 48 h before sorting or left untreated. Cells were sorted into 384-well plates that were acquired from Single Cell Discoveries (Bio-Rad, HSP3801), each well containing 10 µl sterile mineral oil (Sigma-Aldrich, M5310) and 50 nl DNA oligo primer (Sigma-Aldrich, M8410). Cells with increased ploidy were gated according to their Hoechst signal and, as a reference for the whole population, the gate was adjusted in the same sample. After sorting, plates were immediately centrifuged and placed on dry ice. They were stored at -80 °C and were shipped on dry ice to Single Cell Discoveries, where scRNA-seq was performed according to an adapted version of the SORT-seq protocol65 with primers described previously66. Cells were heat-lysed at 65 °C followed by cDNA synthesis. After second-strand cDNA synthesis, all of the barcoded material from one plate was pooled into one library and amplified using in vitro transcription (IVT). After amplification, library preparation was performed according to the CEL-Seq2 protocol67 to prepare a cDNA library for sequencing using TruSeq small RNA primers (Illumina). The DNA library was paired-end sequenced on the Illumina NextSeq 500 system, high output, with the 1 × 75 bp Illumina kit (read 1: 26 cycles, index read: 6 cycles, read 2: 60 cycles).
During sequencing, read 1 was assigned 26 bp and was used to identify the Illumina library barcode, cell barcode and UMI. Read 2 was assigned 60 bp and was used to map to the reference genome Homo sapiens GRCh38.p13 with STARSolo (v.2.7.10b)68. In brief, mapping and generation of count tables were automated using the STARSolo v.2.7.10b and aligner. For mapping, no UMI cut-off was used and intronic reads were not included. Multi-gene reads were not counted. Unsupervised clustering and differential gene expression analysis was performed with the Seurat R toolkit69. For this, logarithmic normalization was applied, which normalizes to the total RNA counts in each cell. This approach consists of dividing each raw UMI count by the total detected RNAs in that cell, multiplying by a scale factor (10,000), adding a pseudocount (1) and performing log transformation. For visualization of the t-SNE, the clustering algorithm used was the original Louvain algorithm where the first 19 principal components were used with a k parameter of 30 and a resolution of 0.5.
Subclustering of the polyploid samples was performed with Louvain resolution of 0.5 for both samples with 50 principal components as identified by principal component analysis. Differential gene expression analysis for all comparisons was done using the Venice method, a nonparametric statistical test for single-cell data70 in BioTuring BBrowser X software (BioTuring). Tables were extracted and plotted in TIBCO Spotfire (v.7.9.1, v.10.10.1). For the AUCellScores the integrated tool in BioTuring BBrowser X was used. For GO-analysis, ShinyGO (v.0.77 and v.0.80) available online (https://bioinformatics.sdstate.edu/go/)71 was used. For generation of Venn diagrams, the tool InteractiVenn (https://www.interactivenn.net)72 was used.
The data analysis for the replicate samples was done as described above, except that a different version of the mapper STARSolo (v.2.7.11b)68 was used. For subclustering of the polyploid cells of the replicate sample, a Louvain resolution of 0.5 was used for the HRAS polyploid sample and a Louvain resolution of 1.0 was used for the Pevo polyploid sample. Sequencing raw data and processed Poisson-corrected data files for both replicates of the polyploidy data set are accessible on GEO (GSE255874).
Cell clustering analysis of the IR-treated samples was performed with the Seurat R Bioconductor package69, using the SC transformed counts generated using the ‘v2’ vst flavour, with Louvain resolution of 0.6 and the first 20 principal components as identified by principal component analysis, for both samples. Dimension reduction was performed using the UMAP method, using the first 20 principal components. Differential expression was performed between treated versus untreated cells using the FindMarkers() function, using the option to return only positive genes. Enriched pathways per cluster were generated using the enrichGO function of the clusterProfiler Bioconductor R package73 or the Enrichr gene list enrichment analysis tool74, using the marker genes identified per cluster from the FindMarkers() function. All R functions were executed on R v.4.4.2 (https://www.R-project.org) and Bioconductor v.3.20. All scRNA-seq and SORT-seq analyses were performed on two independent sets of samples. Sequencing raw data and processed data files for the IR-treated versus UT dataset are accessible on GEO (GSE288487).
Bulk RNA-seq
Cells were treated for 24 h with 4 Gy of IR or left untreated. RNA was extracted from triplicate samples using the TRIzol RNA MiniPrep Plus Kit according to the manufacturer’s protocol. Extracted RNA was prepared for sequencing by the Functional Genomics Center Zurich (FGCZ) using the Illumina TruSeq Total RNA Library Prep assay according to the manufacturer’s protocol. Sequencing was performed on the Illumina NovaSeq 6000 system using the S1 Reagent Kit v1.5 (100 cycles) according to the manufacturer’s protocol. Demultiplexing was performed using the Illumina bcl2fastq Conversion Software (v.2.20.0.422). Individual library sizes ranged from 22 million to 52 million reads.
RNA-seq analysis was performed using the SUSHI framework75, which encompassed the following steps: read quality was inspected using FastQC, and sequencing adaptors were removed using fastp76; pseudoalignment and transcriptomic counts of the RNA-seq reads was performed using the Kallisto Bioconductor R package77 with the GENCODE human genome build GRCh38.p13 (release 37)78; differential expression using the generalized linear model as implemented by the edgeR Bioconductor R package79; and Gene Ontology (GO) term pathway analysis using the hypergeometric over-representation test with the enrichGO function of the clusterProfiler Bioconductor R package73 or the Enrichr gene list enrichment analysis tool74. Additional figures were generated using the exploreDE Shiny app (https://doi.org/10.5281/zenodo.13927692). All R functions were executed on R v.4.4.2 (https://www.R-project.org) and Bioconductor v.3.20. Bulk sequencing data files are accessible on GEO (GSE288485).
Primary antibodies used in this study
H2AX phospho-S139 (BioLegend, 613401, 1:1,000 for IF), pRb (Cell Signaling Technologies, 8516S, 1:500 for IF), p21 (Abcam, ab109520, 1:500 for IF and WB), p53 (Thermo Fisher Scientific, AHO0152, 1:500 for IF and WB), 53BP1 (Novus Biologicals, NB100-304, 1:1,000 for IF), cyclin A (Abcam, ab181591, 1:500 for IF), cyclin E (Abcam, ab208696, 1:1,000 for WB), HRAS (GeneTex, GTX116041, 1:500 for WB), PCNA (Santa Cruz, sc-56, 1:2,000 for WB), tubulin (Sigma-Aldrich, T6199, 1:5,000 for WB), AMBRA1 (Santa Cruz, sc-398204, 1:500 for WB), KAP1 phospho-S824 (Abcam, ab70369, 1:500 for WB), KAP1 (Bethyl, A300-274A, 1:2,000 for WB), CHK1 phospho-S296 (Abcam, ab79758, 1:500 for WB), CHK1 (Abcam, ab40866, 1:500 for WB), RPA32 phospho-S4/8 (Bethyl, A300-245A, 1:1,000 for WB), RPA32/RPA2 (Abcam, ab2175, 1:500 for WB), BrdU (Abcam, ab6326, 1:250 for IF; BD Biosciences, 347580, 1:80 for IF), MCM2 (Santa Cruz, sc-9839, 1:100 for IF) and MCM7 (Santa Cruz, sc-9966, 1:100 for IF).
Secondary antibodies used in this study
Alexa Fluor 647 goat anti-rabbit (Thermo Fisher Scientific, A21244, 1:500 for IF), Alexa Fluor 647 goat anti-mouse (Thermo Fisher Scientific, A21235, 1:500 for IF), Alexa Fluor 568 goat anti-rabbit (Thermo Fisher Scientific, A11036, 1:500 for IF), Alexa Fluor 568 goat anti-mouse (Thermo Fisher Scientific, A11031, 1:500 for IF), Alexa Fluor 555 goat anti-rat (Thermo Fisher Scientific, A21434, 1:250 for IF), Alexa Fluor 488 goat anti-rabbit (Thermo Fisher Scientific, A11034, 1:500 for IF), Alexa Fluor 488 goat anti-mouse (Thermo Fisher Scientific, A11029, 1:500 for IF), Alexa Fluor 488 rabbit anti-goat (Thermo Fisher Scientific, A11078, 1:500 for IF), goat anti-rabbit IgG antibody (H+L), peroxidase (Vector Laboratories, PI-1000-1, 1:10,000 for WB), goat anti-mouse IgG Antibody (H+L), peroxidase (Vector Laboratories, PI-2000-1, 1:10,000 for WB).
Statistics and reproducibility
Apart from time-lapse microscopy, no samples were measured repeatedly, and all other measurements were taken from distinct samples. Statistical analysis was performed in GraphPad Prism (v.9 and 10). Two-tailed unpaired t-tests, χ2 tests, Fisher’s exact tests or one-way ANOVA followed by Tukey’s post hoc test were performed as indicated in the figure legends. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. All extended time-lapse experiments were performed at least twice for each experimental condition (that is, independent biological replicates with different batches of cells and the live-cell microscopy performed in different weeks), each experiment with 1–5 wells per experimental condition (typically 2–3 wells per condition) and multiple images taken per well (typically 9 images per well). Bulk RNA-seq was performed in triplicates, scRNA-seq was performed in duplicates.
Control western blots (Supplementary Figs. 3a,b and 6b) were performed once, control DNA fibre experiments (Extended Data Figs. 9c and 10c) were performed twice. All of the other experiments were performed at least three times, and the presented results were reliably reproduced.
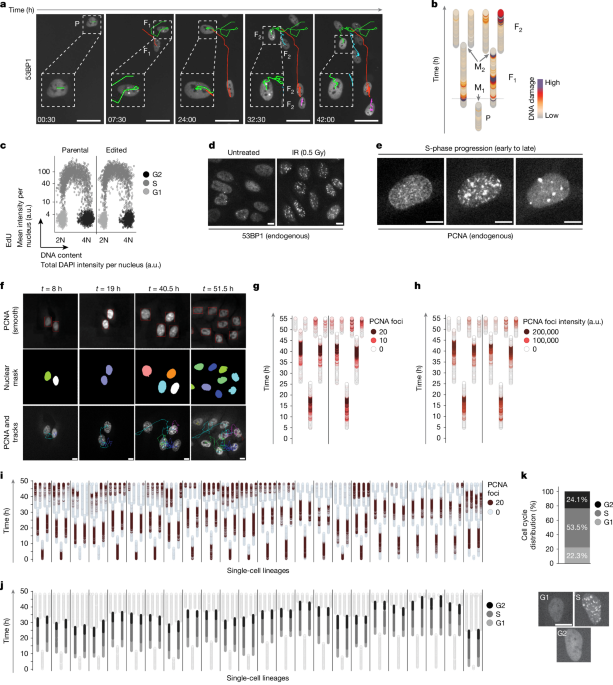
Sample numbers, numbers of experiments performed, statistical tests, exact P values, and definition of error bars and box plots are as follows: Fig. 1c, 3 independent experiments performed, n = 2,955 (parental) and n = 3,018 (edited) cells are shown. Fig. 1d, 3 independent experiments performed, representative images are shown. Fig. 1e, 3 independent experiments performed, representative images are shown. Fig. 1i, 3 independent experiments performed, n = 24 cell lineages shown. Fig. 1j, 3 independent experiments performed, n = 24 lineages shown. Fig. 2a, 3 independent live-cell experiments performed, n = 51 cells analysed; the box plot limits indicate the 25th percentile (Q1) and 75th percentile (Q3); the boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 2c, 3 independent experiments performed, n = 51 (UT), n = 43 (APH) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 2d, 3 independent experiments performed, n = 51 (UT), n = 38 (ATRi) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 3a, P values were derived from a differential gene expression test using the generalized linear model as implemented by edgeR Bioconductor R package with Benjamini–Hochberg multiple test correction, n = 14,664 unique genes. Fig. 3b, P value (two-sided Fisher’s exact test on genes with residuals of >0.5 versus genes with residuals of ≤0.5): 2.45 × 10–12, odds ratio = 1.90, confidence interval (CI): 1.58 to 2.299, n = 9,797 unique genes. Fig. 3c, P values were derived through Enrichr by one-sided right-tailed Fisher’s test with Benjamini–Hochberg multiple-test correction. Fig. 3d, 3 independent experiments performed, n = 5,065 (0 Gy), n = 3,747 (0.25 Gy), n = 3,605 (0.5 Gy), n = 3,632 (1 Gy), n = 4,068 (2 Gy), n = 3,686 (4 Gy), n = 3,938 (10 Gy) cells are shown; the solid line indicates the mean and the dashed lines indicate the s.d. Fig. 3e, 3 independent experiments performed. Fig. 3f, 3 independent experiments performed, n = 855 (UT), n = 1,011 (ATRi), n = 1,205 (IR), n = 965 (ATRi→IR), n = 273 (UT in G1), n = 598 (ATRi in G1), n = 404 (IR in G1), n = 557 (ATRi→IR in G1) cells are shown; P values and CIs γH2AX foci (one-way ANOVA followed by Tukey’s post hoc test): P = 0.0012 (CI: −3.886 to −0.7074) for UT versus ATRi, P < 0.0001 (CI: −5.710 to −2.865) for IR versus ATRi→IR; P values and CIs 53BP1 foci (one-way ANOVA followed by Tukey’s post hoc test): P = 0.0072 (CI: 0.2131 to 1.908) for IR versus ATRi→IR; the solid line indicates the mean and the dashed lines indicate the s.d. Fig. 4a, 3 independent experiments performed, n = 7,347 (control), n = 7,310 (pevonedistat 100 nM), n = 7,215 (pevonedistat 175 nM), n = 7,088 (pevonedistat 250 nM) cells shown. Fig. 4b, 3 independent live-cell experiments performed, n = 557 (G1 peak, control), n = 170 (≥G2 peak, control), n = 221 (G1 peak, pevonedistat), n = 282 (≥G2 peak, pevonedistat) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 4c, 3 independent live-cell experiments performed, n = 557 (G1 peak, control), n = 170 (≥G2 peak, control), n = 221 (G1 peak, pevonedistat), n = 282 (≥G2 peak, pevonedistat) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 4d, 3 independent live-cell experiments performed, n = 557 (G1 peak, control), n = 170 (≥G2 peak, control), n = 221 (G1 peak, pevonedistat), n = 282 (≥G2 peak, pevonedistat) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 4e, 3 independent live-cell experiments performed, representative examples are shown. Fig. 4f, 3 independent live-cell experiments performed, representative images are shown. Fig. 4g, 2 independent live-cell experiments performed, n = 61 (normal), n = 94 (endoreplication), n = 84 (rereplication) cells analysed; where the solid line indicates the mean and the dashed lines indicate the s.d. Fig. 4h, 2 independent live-cell experiments performed, n = 61 (normal), n = 94 (endoreplication), n = 84 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d., P values and CI (two-tailed unpaired t-test): P < 0.0001 (CI: 423.0 to 673.3). Fig. 4i, 2 independent live-cell experiments performed, n = 61 (normal), n = 94 (endoreplication), n = 84 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d.; P values and CI (two-tailed unpaired t-test): P < 0.0001 (CI: 186.4 to 372.6). Fig. 4j, 2 independent live-cell experiments performed, n = 61 (normal), n = 94 (endoreplication), n = 84 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d.; P values and CI (two-tailed unpaired t-test): P < 0.0001 (CI: 738.3 to 2,175). Fig. 4k, 2 independent live-cell experiments performed, n = 61 (normal), n = 94 (endoreplication), n = 84 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d., P values and CI (two-tailed unpaired t-test): P = 0.0022 (CI: 536.2 to 2,404). Fig. 5a, 2 independent live-cell experiments performed, n = 88 (EV), n = 88 (HRAS), n = 489 (G1 peak, EV), n = 191 (≥G2 peak, EV), n = 411 (G1 peak, HRAS), n = 216 (≥G2 peak, HRAS) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Fig. 5f, 3 independent experiments performed, n = 7,347 (control), n = 7,215 (pevonedistat) cells shown. Fig. 5g, 2 independent live-cell experiments performed, n = 20 and 15 (pevonedistat), n = 4 and 5 (etoposide), n = 20 and 20 (RO3306) cells undergoing polyploidization analysed, where solid lines depict the mean and dots represent the cumulative percentage of each experiment.
Extended Data Figures: Extended Data Fig. 1a, 2 independent live-cell experiments performed, representative images are shown. Extended Data Fig. 1c, 1 experiment performed. Extended Data Fig. 1e, 3 independent experiments performed, n = 1,066 (parental UT), n = 1,103 (edited UT), n = 1,133 (parental IR), n = 1,088 (edited IR) cells shown. Extended Data Fig. 1f, 3 independent experiments performed, n = 1,132 (parental UT), n = 1,141 (edited UT), n = 1,123 (parental IR), n = 1103 (edited IR) cells shown. Extended Data Fig. 1g, 2 independent live-cell experiments performed, n = 2,038 (imaged), n = 2,036 (not imaged) cells shown. Extended Data Fig. 1h, 2 independent live-cell experiments performed, n = 2,038 (imaged), n = 2,036 (Not imaged) cells shown. Extended Data Fig. 1i, 2 independent live-cell experiments performed, n = 2,051 (imaged), n = 2,034 (not imaged) cells shown. Extended Data Fig. 2b, 3 independent experiments performed, n = 4,833 (siCtrl UT), n = 4,486 (siCtrl IR), n = 4,510 (si53BP1 UT), n = 4,407 (si53BP1 IR) cells analysed; where the solid line indicates the mean and the dashed lines indicate the s.d.; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 (CI: −0.4840 to −0.4030) for UT siCtrl versus IR siCtrl, P < 0.0001 (CI: 0.5565 to 0.6393) for IR siCtrl versus IR si53BP1. Extended Data Fig. 2d, 3 independent experiments performed, n = 4,453 cells shown. Extended Data Fig. 2e, 3 independent experiments performed, n = 945 (G1), n = 2,963 (S), n = 545 (G2/M) cells shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 (CI: −39.23 to −34.93) for G1 versus S, P < 0.0001 (CI: 31.86 to 37.23) for S versus G2/M; box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 3b, 3 independent live-cell experiments performed, n = 23 lineages shown. Extended Data Fig. 3d, 3 independent experiments performed, representative results are shown. Extended Data Fig. 3e, 3 independent live-cell experiments performed, n = 26 lineages shown. Extended Data Fig. 3g, 3 independent live-cell experiments performed, n = 27 lineages shown. Extended Data Fig. 4b, 3 independent experiments performed, representative images are shown. Extended Data Fig. 4c, 3 independent experiments performed, n = 1,659 (control), n = 1,324 (replication stress) cells shown. Extended Data Fig. 4d, 3 independent experiments performed, n = 1,659 (control), n = 1,324 (replication stress) cells shown. Extended Data Fig. 4e, 3 independent experiments performed, n = 1,659 (control), n = 1,324 (replication stress) cells shown. Extended Data Fig. 4f, 3 independent experiments performed, n = 4,835 (untreated), n = 2,781 (APH), n = 3,217 (ATRi) cells shown. Extended Data Fig. 4g, 3 independent experiments performed, n = 3,022 (untreated), n = 2,966 (APH), n = 3,008 (ATRi) cells shown. Extended Data Fig. 4h, 3 independent experiments performed, n = 1,269 (untreated), n = 1,183 (APH), n = 1,053 (ATRi) cells shown. Extended Data Fig. 4i, 3 independent experiments performed, n = 3,165 (untreated), n = 2,932 (APH), n = 3,054 (ATRi) cells shown. Extended Data Fig. 5a, 3 independent live-cell experiments performed, n = 51 (UT), n = 43 (APH) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 5b, 3 independent live-cell experiments performed, n = 51 (UT), n = 38 (ATRi) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 5c, 3 independent live-cell experiments performed, n = 51 (UT), n = 43 (APH) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 5d, 3 independent live-cell experiments performed, n = 51 (UT), n = 38 (ATRi) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 5e, 3 independent live-cell experiments performed, n = 29 (53BP1 foci) and n = 27 (γH2AX, p53, p21) sister cell pairs for UT, n = 49 (53BP1 foci), n = 33 (γH2AX), and n = 31 (p53, p21) sister cell pairs for APH, n = 36 (53BP1 foci) and n = 31 (γH2AX, p53, p21) sister cell pairs for ATRi; P values (χ2 test): P = 0.0004 for UT versus ATRi (53BP1 foci), P = 0.0012 for UT versus ATRi (γH2AX levels), P = 0.0284 for UT versus APH (p53 levels), P = 0.0031 for UT versus ATRi (p53 levels), P = 0.0131 for UT versus APH (p21 levels), P < 0.0001 for UT versus ATRi (p21 levels). Extended Data Fig. 6b, 3 independent experiments performed, n = 3,592 (siCtrl UT), n = 4,824 (siCtrl IR), n = 3,267 (si53BP1 UT), n = 4,662 (si53BP1 IR) cells shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 for UT siCtrl versus IR siCtrl (CI: −1.431 to −1.288), P < 0.0001 for IR siCtrl versus IR si53BP1 (CI: 1.413 to 1.547); the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 6d, 3 independent experiments performed, n = 4,083 cells shown. Extended Data Fig. 6e, 3 independent experiments performed, n = 1,972 (G1), n = 1,759 (S), n = 352 (G2) cells shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 for G1 versus S (CI: −38.77 to −36.22), P < 0.0001 for S versus G2 (CI: 31.77 to 36.31); box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 6f, 3 independent experiments performed, n = 2,519 (untreated), n = 2,508 (APH), n = 2,501 (ATRi) cells shown. Extended Data Fig. 6g, 3 independent experiments performed, n = 2,519 (untreated), n = 2,508 (APH), n = 2,501 (ATRi) cells shown. Extended Data Fig. 6h, 3 independent experiments performed, n = 2,267 (untreated), n = 2,134 (APH), n = 2,143 (ATRi) cells shown. Extended Data Fig. 6i, 3 independent experiments, n = 2,267 (untreated), n = 2,134 (APH), n = 2,143 (ATRi). Extended Data Fig. 6j, 3 independent experiments performed, representative results are shown. Extended Data Fig. 7b, P values were derived through Enrichr using a one-sided right-tailed Fisher’s test with Benjamini–Hochberg multiple-test correction. Extended Data Fig. 7c, P values were derived from differential gene expression test using the generalized linear model as implemented by the edgeR Bioconductor R package with Benjamini–Hochberg multiple-test correction, boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. n = 3 independent samples. Extended Data Fig. 7d, boxes represent the IQR with the median value (solid lines), the whiskers define the lower and upper adjacent value, and the violin plot represents the total distribution. n = 346 (UT), n = 227 (IR) cells analysed. Extended Data Fig. 7e, boxes represent the IQR with the median value (solid lines), the whiskers define the lower and upper adjacent value, and the violin plot represents the total distribution. n = 346 (UT), n = 227 (IR) cells for experiment 1 and n = 355 (UT), n = 230 (IR) cells for experiment 2. Extended Data Fig. 7f, n = 573 cells analysed. Extended Data Fig. 7g, n = 346 (UT), n = 227 (IR) cells analysed. Extended Data Fig. 7h, P values were derived by one-sided right-tailed Fisher’s test with Benjamini–Hochberg multiple-test correction through clusterProfiler. Extended Data Fig. 7i, Pearson’s correlation coefficient, n = 9,279 all genes, n = 418 DDR genes. Extended Data Fig. 8a, 3 independent live-cell experiments performed. Extended Data Fig. 8b, 3 independent live-cell experiments performed. Extended Data Fig. 8c, 3 independent experiments performed, n = 423 (control), n = 158 (IR) cells shown; P values and CI (two-tailed unpaired t-test): P < 0.0001 (CI: 3.246 to 3.978); the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 8d, 3 independent experiments performed, n = 4,049 (UT), n = 4,079 (4-OHT), n = 4,377 (4-OHT-IAA) cells shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 for UT versus 4-OHT (CI: −2.414 to −2.093), P < 0.0001 for 4-OHT versus 4-OHT-IAA (CI: 1.669 to 1.984); the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 8e, 3 independent experiments performed, n = 4,049 (UT), n = 4,079 (4-OHT), n = 4,377 (4-OHT-IAA) cells shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 for UT versus 4-OHT (CI: −6.751 to −5.977), P < 0.0001 for 4-OHT versus 4-OHT-IAA (CI: 2.990 to 3.749); the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 8f, 2 independent live-cell experiments performed. Extended Data Fig. 8g, 3 independent experiments performed, n = 621 (−4-OHT), n = 639 (+4-OHT + IAA) cells shown; P values and CI (two-tailed unpaired t-test): P = 0.0003 (CI: 0.1963 to 0.6682); the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 8h, 3 independent experiments performed, n = 2,428 (WT control), n = 614 (WT IR), n = 2,406 (p53KO control), n = 2,106 (p53-KO IR) cells shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 for WT control versus WT IR (CI: −0.5428 to −0.2159), P < 0.0001 for p53KO control versus p53KO IR (CI: −0.9969 to −0.7810); the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 8i, 2 independent live-cell experiments performed, n = 32 (53BP1 foci) and n = 52 (γH2AX, p53, pRb) sister cell pairs for UT; n = 54 (53BP1 foci) and n = 68 (γH2AX, p53, pRb) sister cell pairs for 4 Gy; P values (χ2 test): P = 0.0088 for UT versus 4 Gy (53BP1 foci), P = 0.0080 for UT versus 4 Gy (γH2AX levels), P = 0.0397 for UT versus 4 Gy (p53 levels), P = 0.0176 for UT versus 4 Gy (pRb levels). Extended Data Fig. 8j, 2 independent live-cell experiments performed. Extended Data Fig. 9a, 3 independent experiments performed, n = 45,070 (control), n = 16,136 (pevonedistat 100 nM), n = 10,579 (pevonedistat 175 nM), n = 7,551 (pevonedistat 250 nM) cells shown. Extended Data Fig. 9b, 3 independent experiments performed, representative images shown. Extended Data Fig. 9c, 2 independent experiments performed, DNA fibre length from n = 100 (control), n = 100 (pevonedistat) fibres shown; P values and CI (two-tailed unpaired t-test): P < 0.0001 (CI: −11.40 to −9.096); the red solid line indicates the median. Extended Data Fig. 9d, 3 independent experiments performed, n = 2,142 (control), n = 2,120 (pevonedistat) cells shown. Extended Data Fig. 9e, 3 independent experiments performed, n = 1,368 (control), n = 1,340 (pevonedistat) cells shown. Extended Data Fig. 9f, 3 independent live-cell experiments performed, representative examples are shown. Extended Data Fig. 9g, 3 independent live-cell experiments performed, representative examples are shown. Extended Data Fig. 9h, 3 independent live-cell experiments performed, n = 9, n = 13, n = 15 (G1), n = 8, n = 12, n = 15 (S) cells undergoing polyploidization; data are mean ± s.d. Extended Data Fig. 9i, 2 independent live-cell experiments performed, representative examples are shown. Extended Data Fig. 9j, 2 independent live-cell experiments performed, representative examples are shown. Extended Data Fig. 9k, 2 independent experiments performed, n = 2,221 (siCtrl), n = 2,197 (siGMNN) cells shown. Extended Data Fig. 9l, 2 independent live-cell experiments performed, n = 43 (normal), n = 15 (endoreplication), n = 37 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 9m, 2 independent live-cell experiments performed, n = 43 (normal), n = 15 (endoreplication), n = 37 (rereplication) cells analysed; P values and CI (two-tailed unpaired t-test): P = 0.0256 (CI: 6.720 to 98.88) for endoreplication versus rereplication; the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 9n, 2 independent live-cell experiments performed, n = 43 (normal), n = 15 (endoreplication), n = 37 (rereplication) cells analysed; P values and CI (two-tailed unpaired t-test): P = 0.0214 (CI: 4.769 to 56.93) for endoreplication versus rereplication; the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 9o, 2 independent live-cell experiments performed, n = 43 (normal), n = 15 (endoreplication), n = 37 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 9p, 2 independent live-cell experiments performed, n = 43 (normal), n = 15 (endoreplication), n = 37 (rereplication) cells analysed; the solid line indicates the mean and the dashed lines indicate the s.d. Extended Data Fig. 10a, 3 independent experiments performed, representative results are shown. Extended Data Fig. 10b, 3 independent experiments performed, representative results are shown. Extended Data Fig. 10c, 2 independent experiments performed, DNA fibre length of n = 120 (EV), n = 120 (HRAS), n = 120 (cyclin E) fibres shown; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 (CI: 1.699 to 5.797) for EV versus HRAS, P = 0.0003 (CI: 1.399 to 5.497) for EV versus cyclin E; the red solid line indicates the median. Extended Data Fig. 10d, 4 independent experiments performed, n = 100 cells per replicate (EV), n = 100 cells per replicate (HRAS), n = 100 cells per replicate (cyclin E); P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P = 0.0234 (CI: −25.86 to −3.645) for EV versus HRAS, P = 0.0078 (CI: −17.75 to −5.749) for EV versus cyclin E; data are mean ± s.d. Extended Data Fig. 10e, 2 independent live-cell experiments performed, n = 88 (EV), n = 489 (G1 peak, EV), n = 191 (≥G2 peak, EV) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 10f, 2 independent live-cell experiments performed, n = 88 (EV), n = 88 (HRAS), n = 489 (G1 peak, EV), n = 191 (≥G2 peak, EV), n = 411 (G1 peak, HRAS), n = 216 (≥G2 peak, HRAS) cells analysed; box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 10g, 2 independent live-cell experiments performed, n = 88 (EV), n = 92 (cyclin E), n = 489 (G1 peak, EV), n = 191 (≥G2 peak, EV), n = 548 (G1 peak, cyclin E), n = 248 (≥G2 peak, cyclin E) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 10h, 2 independent live-cell experiments performed, n = 88 (EV), n = 92 (cyclin E), n = 489 (G1 peak, EV), n = 191 (≥G2 peak, EV), n = 548 (G1, cyclin E), n = 248 (≥G2 peak, cyclin E) cells analysed; box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 11a, 3 independent experiments performed, n = 3,097 (empty vector UT), n = 3,026 (HRAS UT), n = 3,144 (cyclin E UT), 3,082 (empty vector IR), n = 3,149 (HRAS IR), n = 3,121 (cyclin E IR) cells shown. Extended Data Fig. 11b, 3 independent experiments performed, representative images are shown. Extended Data Fig. 11c, 3 independent experiments with multiple replicate samples performed, n = 3,093, n = 3,076, n = 3,045, n = 3,019, n = 3,020, n = 3,031 (empty vector UT), n = 3,026, n = 3,073, n = 3,126, n = 3,064, n = 2,994, n = 3,043 (HRAS UT), n = 3,144, n = 3,012, n = 3,003, n = 3,013, n = 2,991, n = 3,090 (cyclin E UT), n = 3,082, n = 3,044, n = 3,014, n = 3,003, n = 3,068, n = 2,985 (empty vector IR), n = 3,149, n = 3,091, n = 3,051, n = 3,035, n = 3,075, n = 2,907 (HRAS IR), n = 3,121, n = 2,869, n = 3,036, n = 3,083, n = 2,467, n = 2,179 (cyclin E IR) cells analysed; P values and CIs (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 (CI: −2.608 to −0.9585) for empty vector UT versus HRAS UT, P < 0.0001 (CI: −2.291 to −0.6419) for empty vector UT versus cyclin E UT, P < 0.0001 (CI: −2.575 to −0.9252) for empty vector UT versus empty vector IR, P < 0.0001 (CI: −4.891 to −3.242) for empty vector UT versus HRAS IR, P < 0.0001 (CI: −3.908 to −2.259) for empty vector UT versus cyclin E IR; data are mean ± s.d. Extended Data Fig. 11d, 3 independent experiments performed, n = 15,249 (empty vector UT), n = 15,262 (HRAS UT), n = 15,144 (cyclin E UT), n = 15,200 (empty vector IR), n = 15,378 (HRAS IR), n = 14,548 (cyclin E IR) cells shown. Extended Data Fig. 11f, 3 independent live-cell experiments performed, representative examples are shown. Extended Data Fig. 11g, 2 independent experiments performed, representative results are shown. Extended Data Fig. 11h, 3 independent experiments performed, n = 4,867 (−Dox), n = 4,995 (+Dox) cells shown. Extended Data Fig. 12a, 2 independent live-cell experiments performed, n = 12 (−Dox), n = 414 (G1 peak, −Dox), n = 271 (≥G2 peak, −Dox) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 12b, 2 independent live-cell experiments performed, n = 12 (−Dox), n = 36 (+Dox), n = 414 (G1 peak, −Dox), n = 271 (≥G2 peak, −Dox), n = 286 (G1 peak, +Dox), n = 259 (≥G2 peak, +Dox) cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Extended Data Fig. 12c, 2 independent live-cell experiments performed, n = 174 (experiment 1), n = 231 (experiment 2), mean (solid lines) and individual percentage values are depicted.
Supplementary Figures: Supplementary Fig. 2a, 3 independent experiments performed, n = 10,096 (unchallenged), n = 3,319 (APH), n = 3,388 (ATRi) cells shown. Supplementary Fig. 2b, 3 independent experiments performed, n = 10,096 (unchallenged), n = 3,319 (APH), n = 3,388 (ATRi) cells shown. Supplementary Fig. 2c, 3 independent experiments performed, n = 1,021 (G1 unchallenged), n = 1,764 (S unchallenged), n = 604 (G2/M unchallenged), n = 510 (G1 APH), n = 2,268 (S APH), n = 541 (G2/M APH), n = 1,770 (G1 ATRi), n = 899 (S ATRi), n = 719 (G2/M ATRi) cells shown. Supplementary Fig. 2d, 3 independent experiments performed, representative images are shown. Supplementary Fig. 2e, 3 independent live-cell experiments performed, n = 10 lineages per condition; P values and CIs for perturbed replication (one-way ANOVA followed by Tukey’s post hoc test): P < 0.0001 (CI: −2.148 to −0.6516) for UT versus APH, P < 0.0001 (CI: −2.648 to −1.152) for UT versus ATRi, P < 0.0001 (CI: −2.148 to −0.6516) for UT versus IR; P values and CIs for >10 53BP1 foci (one-way ANOVA followed by Tukey’s post hoc test): P = 0.0003 (CI: −1.582 to −0.4182) for UT versus IR; P values and CIs for number of divisions (one-way ANOVA followed by Tukey’s post hoc test): P = 0.0002 (CI: 0.6531 to 2.347) for UT versus APH, P < 0.0001 (CI: 0.7531 to 2.447) for UT versus ATRi, P = 0.0011 (CI: 0.4531 to 2.147) for UT versus IR. Supplementary Fig. 3a, 1 experiment performed. Supplementary Fig. 3b, 1 experiment performed. Supplementary Fig. 3c, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3d, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3e, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3f, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3g, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3h, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3i, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3j, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 3k, 2 independent live-cell experiments performed; an example cell lineage is shown. Supplementary Fig. 4a, 2 independent live-cell experiments performed, n = 21 lineages shown. Supplementary Fig. 4b, 2 independent live-cell experiments performed, n = 17 lineages shown. Supplementary Fig. 4c, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 4d, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 4e, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 4f, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 4g, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 4h, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 4i, 2 independent live-cell experiments performed, n = 20 lineages shown. Supplementary Fig. 5a, 2 independent live-cell experiments performed, n = 27 cells analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Supplementary Fig. 5b, 2 independent live-cell experiments performed, n = 27 cells (siCtrl), n = 47 cells (siAMBRA1) analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Supplementary Fig. 5c, 2 independent live-cell experiments performed, n = 27 cells (siCtrl), n = 47 cells (siAMBRA1), n = 38 cells (siAMBRA1 + APH) analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Supplementary Fig. 5d, 2 independent live-cell experiments performed, n = 27 cells (siCtrl), n = 47 cells (siAMBRA1), n = 25 cells (siAMBRA1 + ATRi) analysed; the box plot limits indicate Q1 and Q3; boxes represent the IQR with the median value (solid lines); the whiskers define the lower and upper adjacent value; dots show outliers smaller than Q1 − 1.5 × IQR and greater than Q3 + 1.5 × IQR. Supplementary Fig. 6a, 3 independent experiments performed; P values and CI (two-tailed unpaired t-test): P = 0.0006 (CI: 17.60 to 31.07) for 0.1 μM ATRi, P = 0.0114 (CI: 1.740 to 7.593) for 0.2 μM ATRi, P = 0.0111 (CI: 1.264 to 5.403) for 0.3 μM ATRi; data are mean ± s.d. Supplementary Fig. 6b, 1 experiment performed. Supplementary Fig. 7b, 2 independent live-cell experiments performed, an example lineage is shown. Supplementary Fig. 7c, 2 independent live-cell experiments performed, an example lineage is shown. Supplementary Fig. 7d, 2 independent live-cell experiments performed, an example lineage is shown. Supplementary Fig. 7e, 2 independent live-cell experiments performed, an example lineage is shown. Supplementary Fig. 8a, 3 independent experiments performed, n = 1,482 (control), n = 1,511 (IR) cells shown. Supplementary Fig. 8b, 2 independent live-cell experiments performed, n = 60 sister cell pairs for UT, n = 60 sister cell pairs for IR; P value (Fisher’s exact test): P = 0.0013. Supplementary Fig. 12i, 3 independent experiments performed, n = 2,240 (control), n = 1,927 (etoposide), n = 1,993 (RO3306) cells shown. Supplementary Fig. 12j, 3 independent experiments performed, n = 4,356 (control), n = 3,087 (etoposide), n = 3,955 (RO3306) cells shown. Supplementary Fig. 12k, 3 independent experiments performed, n = 4,356 (control), n = 3,087 (etoposide), n = 3,955 (RO3306) cells shown. Supplementary Fig. 12l, 3 independent experiments performed, n = 1,503 (G1/early S control), n = 1156 (mid-S control), n = 637 (late S/G2 control), n = 516 (G1/early S etoposide), n = 872 (mid-S etoposide), n = 1,801 (late S/G2 etoposide), n = 981 (G1/early S RO3306), n = 630 (mid-S RO3306), n = 1,249 (late S/G2 RO3306) cells shown.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.