Chemicals and antibodies
The antibodies and chemicals used in this study with their details are listed in Supplementary Table 10.
Plasmids
Plasmids (wild-type PLA2G15-6xHis and PLA2G15-6xHis S198A) were obtained from a previous study, with Addgene IDs 213603 and 213605, respectively7.
Animal studies
Mice were acquired and maintained, as previously described17. The mice were housed in a controlled environment with regulated 20–26 °C temperature and 30–70% relative humidity, 12-h light/dark cycles and access to food and water. Supplies were checked daily, and the cages were cleaned every 4–5 days. All mouse procedures were conducted in accordance with the approved guidelines of the Administrative Panel on Laboratory Animal Care at Stanford University. The LysoTag mouse was previously reported (Jackson Laboratory (JAX); strain no. 035401)17. GbaD409V was obtained from JAX (strain no. 019106)53. PLA2G15-deficient mice were obtained, housed and used in accordance with approved Institutional Animal Care and Use Committee protocols at the University of Wisconsin–Madison52,54. Before euthanasia and necropsy, the PLA2G15-deficient mice were fasted for 6 h.
Mouse genotypes were confirmed by pPCR using DirectPCR (Thermo Fisher Scientific; AA4647500) according to the manufacturer’s guidelines. Briefly, tail clippings were incubated in DirectPCR overnight at 60 °C for 16 h, followed by the addition of proteinase K and further incubation for 1 h. PCR was performed with PLA2G15 primers (Pla2g15_1: 5′- GAATTCCTAGACCCCAGCAAGAGGAATGTG -3′; Pla2g15_2:5′-ACTGCTCCCCCTCCCCAGAGATGGATATTT-3′) using a program of 4 min at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 55 °C, 1 min at 72 °C and finally with an 8-min incubation at 72 °C. The results of genotyping were validated using reverse transcription PCR (RT–PCR) and western blotting of the excised tissue. These mice were used in experiments, as shown in Fig. 3. For all animal studies, the samples were randomized and blinded when possible, with all sample sizes, sexes and ages detailed in the legends and/or related supplementary tables.
Generation of Pla2g15
−/− mice for the NPC1 rescue experiment
Constitutive Pla2g15−/−(TAC) mice were obtained through CRISPR–Cas9-mediated gene editing at Taconic Biosciences in accordance with national and international regulations. Animal procedures were evaluated by veterinarians and Taconic’s Institutional Animal Care and Use Committee or an equivalent oversight body to ensure the elimination or minimization of the potential for pain or distress. The gene targeting strategy was on the basis of the National Center for Biotechnology Information (NCBI) transcript NM_133792.3 (Pla2g15) and was designed to delete exon 3 from the transcript (proximal single guide RNA (sgRNA): 5′-TCGATGCCATTCATGGACTT-3′ and distal sgRNA: 5′-GGCCTACTACGTTGCTTTGA-3′), resulting in the loss of function of PLA2G15 by generating a frameshift from exon 2 to all downstream exons and creating a premature stop codon in exon 4.
Pla2g15 exon 3 flanking primers (forward: 5′-ACTCACTTTATAGACCAGGTTGGC-3′ and reverse: 5′- CAGAGACAGTGAACCTAAGGGC-3′) were designed to amplify and confirm both wild-type and edited Pla2g15 alleles.
In brief, Cas9 protein and sgRNAs were injected into BALB/c zygotes, and G0 animals were genotyped by PCR analysis, as described above. Founders were identified and used for further breeding activities to create heterozygous Pla2g15−/−(TAC) mice. These mice were subsequently used to generate homozygous Pla2g15−/−(TAC) mice, which were used in experiments, as shown in Fig. 5.
Lifespan analysis and neurological phenotyping
Pla2g15−/−(TAC) BALB/c mice were crossed with Npc1m1N/J heterozygous animals (JAX no. 003092), and from the resulting offspring, double-HET mice were used to obtain the following genotypes (gene order: Npc1m1N/J/Pla2g15): wild type/wild type, homozygous/wild type, homozygous/heterozygous, homozygous/knockout and wild type/knockout. Wild type represents Npc1+/+ or Pla2g15+/+, knockout represents Npc1+/+Pla2g15−/−, HOM represents Npc1−/−Pla2g15+/+ and heterozygous represents Pla2g15+/−. In total, 60 animals were included in the phenotypic analysis, with 12 mice of each of the different genotypes (six male and six female mice). The body weights of all mice were assessed every other day starting at 3 weeks of age after the pups were weaned. Motor coordination was assessed weekly using the rotarod test from 6 to 9 weeks of age. The neurological composite phenotype score, consisting of the ledge test, hindlimb clasping, gait, kyphosis, tremor and grooming scores, was performed on a weekly basis starting at 6 weeks of age. These tests were performed according to an established protocol55. A higher composite score indicates worse neurological performance and was calculated using hindlimb clasping, grooming, tremor, gait, kyphosis and ledge test. Starting at 8 weeks of age, all animals carrying the Npc1m1N/J HOM genotype were subjected to daily monitoring of clinical signs or termination criteria scoring, along with special care (such as providing wet food), which exceeded the standard health checks and animal care. Experiments were performed at QPS Austria in compliance with the Animal Care and Welfare Committee and animal welfare regulations from local authorities.
CSF and plasma analysis
At 8 weeks of age (postnatal day 56 ± 2 days), the mice were terminally anaesthetized by intraperitoneal injection of pentobarbital (600 mg kg−1), and CSF and blood plasma were obtained. The NfL levels were assessed using ELISA 10-7001 CE from UmanDiagnostics in plasma and CSF. Plasma samples were diluted 1:3 and CSF 1:30 (wherever possible) in the assay buffer and analysed according to the manufacturer’s protocol.
AST and ALT concentrations were measured using commercial kits (catalogue nos. 04467493190 and 04467388190, respectively; Roche), according to the International Federation of Clinical Chemistry and Laboratory Medicine with pyridoxal phosphate activation (Roche). Therefore, kinetic measurement of the enzyme activity with a redox reaction of nicotinamide adenine dinucleotide hydrogen was performed using l-aspartate and 2-oxoglutarate as substrates for AST measurement and l-alanine and 2-oxoglutarate as substrates for ALT measurement. The concentrations of the two enzymes were measured using a Roche cobas 6000/c501 analyser.
Histoprocessing
The CNS, livers, spleens and lungs of all animals were sampled at necropsy, frozen and transferred to AnaPath Services GmbH for histoprocessing. The samples were placed in formalin, embedded in paraffin wax and cut to a nominal thickness of 4 µm. The livers, spleens and lungs were stained with haematoxylin and eosin. In the CNS, five sagittal serial sections of the brain were performed. One section was stained with haematoxylin and eosin, and four sections were used for immunohistochemistry and labelled with antibodies against calbindin (ab229915; lot no. GR3361538-6; Abcam) for targeting Purkinje cells, myelin basic protein (ab218011; lot no. 1007654-30; Abcam) for targeting myelin sheaths, IBA1 (ab178846; lot no. 1002201-50; Abcam) for targeting microglia and macrophages and glial fibrillary acidic protein (ab7260; lot no. GR3454901-1; Abcam) for targeting astrocytes. Immunohistochemistry was performed using a BOND-III (Leica Biosystems).
Histopathological evaluation
Histopathological evaluation was performed by an EBVS European Veterinary Specialist in Pathology using an Olympus BX46 microscope. Illustrative microscopic images were captured with an Olympus SC50 camera. Evaluation of the CNS was performed independently in the following neuroanatomical locations: cerebellum, hindbrain (pons and medulla), midbrain, interbrain (thalamus and hypothalamus), basal ganglia (striatum and pallidum), hippocampus, isocortex (somatomotor, somatosensory and visual areas) and olfactory bulb. Histological changes were described according to distribution, severity and morphological characteristics. Severity scores were assigned on a scale of 1–5 as follows: grade 1, minimal; grade 2, slight; grade 3, moderate; grade 4, marked; grade 5, severe. Histopathological findings recorded in different neuroanatomical locations of the CNS included the following: Purkinje cell loss (decreased number of soma/decreased dendrites), microgliosis (increased number/activation of microglia), astrocytosis (increased number/activation of astrocytes), demyelination and foam cell formation. After independent evaluation of the aforementioned neuroanatomical regions, the mean severity score for each finding throughout the CNS was calculated. Histopathological findings in other organs included the following: hyperplasia of Kupffer cells (increased number/activation/foamy cytoplasm) and vacuolation of hepatocytes (cytoplasmic alteration/glycogen/lipid accumulation) in the livers, hyperplasia of histiocytes (increased number/activation/foamy cytoplasm) and atrophy of lymphocytes (decreased number in the white pulp) in the spleens, and alveolar macrophages (activated/foamy cytoplasm) and cell debris in the alveolar lumen of the lungs.
Histomorphometry
Histomorphometric analyses were performed on calbindin-immunolabelled CNS sections in all the animals to quantify the number of Purkinje cells. Slides were scanned using an Olympus Slideview VS200 slide scanner coupled to an Olympus VS-264C camera using a ×40 objective to obtain whole slide images (WSIs). Histomorphometric evaluation was conducted in QuPath56, quantitative pathology and bioimage analysis software, v.0.4.3. The cerebellum was annotated as the region of interest (ROI). A cell detection algorithm was used to detect the soma of calbindin-positive cells (that is, Purkinje cells).
Cell culture
HEK293T cells (American Type Culture Collection CRL-3216), HeLa cells (American Type Culture Collection CRM-CCL2) and their derivatives were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific) with 2 mM glutamine, 100 IU ml−1 penicillin and 100 µg ml−1 streptomycin (Thermo Fisher Scientific). HEK293T cells were authenticated using short tandem repeat profiling. The cell cultures were maintained at 37 °C in an incubator with 5% CO2. PLA2G15-deficient clone1 cells with LysoTag were obtained from a previous study7. For protein expression, Expi293F cells (Thermo Fisher Scientific) were maintained in a shaking incubator at 37 °C with 8% CO2. These cells were routinely tested for mycoplasma contamination.
Generation of knockout cell lines using CRISPR–Cas9 technology
PLA2G15 knockout clone1 cells have been described previously7. PLA2G15 knockout HEK293T clone2 and clone3 and HeLa cells were generated following the Synthego Gene Knockout Kit v.2 protocol using the following mixed guide sgRNA sequences: GGGGCGGAGGUGGAGGCCCA; UUAGCAGCAGCAAGAGGAAC and CCUCACCCAGCACCACUGGG.
In short, a combined 180-pmol sgRNA and 20-pmol Cas9 (Integrated DNA Technologies) was incubated at room temperature for 10 min. Concurrently, HEK293Ts were washed with PBS, detached using enzyme-free dissociation buffer (Thermo Fisher Scientific) and counted to determine their density. A suspension of 150,000 cells was added to the newly formed ribonucleoprotein complexes, transferred to a Lonza Nucleocuvette and electroporated using the CM-130 program code. Following electroporation, the cells were placed in growth medium and transferred to a 24-well plate. Single-cell populations were obtained using limited dilution, and the sgRNA target sites were sequenced after PCR amplification. Finally, genetic knockouts were confirmed using Synthego ICE CRISPR analysis, an insertion and deletion deconvolution web-based tool (https://synthego.com).
Cell volume measurement
Beckman Z2 particle counter and size analyser was used to determine cell count and volume. The filtering criteria were configured to select cells between 10 and 30 μm in size.
Lysosomal immunopurification from mouse tissues and HEK293Ts
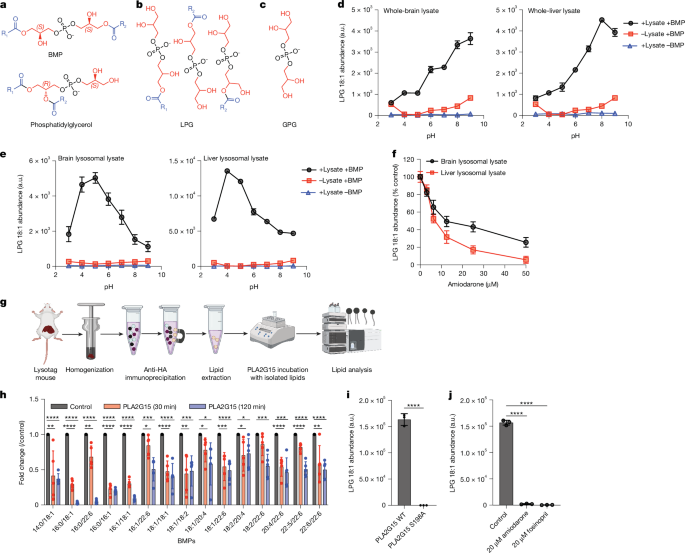
Lysosomes were purified from the brains and livers of mice and HEK293T cells, following established protocols14,17. The brains of mice were obtained after euthanasia. Small uniform liver samples were collected using a 4-mm-diameter biopsy punch to minimize variability between samples. Both brain and liver tissues were used immediately without freezing using the steps described in the lysosomal immunopurification (LysoIP) protocols14,17. During the lysosomal lysate enzyme assays, whole brain or liver tissue was sectioned into three pieces and dounced 30 times with 3-ml PBS supplemented with protease inhibitor. The resulting lysates were centrifuged at 1,000g at 4 °C for 10 min. The supernatant was then transferred to a 15-ml Falcon tube containing 0.5-ml anti-haemagglutinin beads and incubated at 4 °C for 1 h with rotational mixing. Subsequently, the mixture was thoroughly washed and isolated to obtain protein fractions suitable for lysosomal lysate experiments, as previously described7. Before enzymatic assays were performed, the protein concentrations in the lysates were quantified using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). For cell-based assay, an equal number of cells were cultured and allowed to reach at least 80% confluence before each LysoIP. The homogenization and LysoIP from HEK293Ts were adapted from a previous study14 and (https://doi.org/10.17504/protocols.io.bybjpskn).
Expression and purification of proteins
Expi293F cells were cultured using a combination of two-thirds FreeStyle 293 and one-third Expi293 media and transfected with the PLA2G15 plasmids at a density of 3E6 cells ml−1. Using FectoPRO transfection reagent (Polyplus), transfection was performed at a ratio of 1.3-µl FectoPRO to 0.5-µg plasmid DNA per millilitre of cells. Following transfection, glucose and valproic acid were added immediately to reach the final concentrations of 0.4% glucose (w/v) and 0.05% valproic acid (w/v). Equivalent amounts of valproic acid and glucose were added 1 day after transfection, and cells were harvested 3 days after transfection. The culture medium was harvested and combined with PBS in a 1:1 ratio. HisPur Ni-NTA resin was added to the mixture and incubated at 4 °C for 16 h. The resin was washed four times consecutively using different wash compositions before being packed onto a column. The first wash included 50 mM HEPES (pH 7.25), 500 mM NaCl, 0.1 mM EDTA, 5 mM β-mercaptoethanol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20 mM imidazole (pH 7.25), cOmplete EDTA-free protease inhibitor cocktail (Roche), 5% glycerol (v/v) and 1% Triton X-100 (v/v). The second wash was identical to the first wash, except for the addition of 10 mM imidazole (pH 7.25) and the absence of Triton X-100 and protease inhibitor. Imidazole was absent from the third wash, which was identical to the second wash except that it contained 250 mM NaCl. The fourth wash was identical to the third wash, except for the addition of 125 mM NaCl. Buffer containing 300 mM imidazole (pH 7.25), 50 mM HEPES (pH 7.25), 125 mM NaCl, 5% glycerol (v/v), 5 mM β-mercaptoethanol, 1 mM dithiothreitol, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and protease inhibitors was used to elute the protein. Using an Amicon 10-kDa MWCO concentrator, the eluted protein was concentrated and further purified by size exclusion chromatography on a Superdex 200 10/300 column. The fractions were then combined, concentrated using an Amicon 10-kDa MWCO concentrator to achieve a final protein concentration of 1 mg ml−1 and swiftly frozen in liquid nitrogen.
Preparation of liposomes
Unless otherwise specified, 18:1 Diether PC (Avanti) was used to make lipid liposomes. The lipids were dried and combined in a tube with 2-ml water and 3-ml diisopropyl ether to create liposomes that contained only glycerophospholipids. The resulting mixture was sonicated for 10 min in a water bath to create tiny unilamellar vesicles, which were subsequently dried. Dried liposomes were resuspended in water, and Stewart assay was used to determine their concentration.
Enzyme assays
Recombinant PLA2G15-6xHis at 100 nM concentration was mixed with phospholipid substrates (BMP or phosphatidylglycerol isomers) of varying concentrations in 50 mM sodium acetate:acetic acid at pH 4.5 and 150 mM NaCl. Liposomes containing phospholipids had an 18:1 Diether PC that was non-cleavable. All lipids and their catalogue numbers are provided in Supplementary Table 10. The 2,2′ R,R BMP and 2,2′ S,R BMP compounds were obtained from WuXi AppTec. The mixture was incubated at 37 °C for 30 s to monitor products and for 10 min to monitor substrates, followed by heat inactivation at 95 °C for 5 min. To adjust for background in each experiment, controls containing all components except the enzyme were prepared for each substrate. A time course was carried out at various substrate concentrations to guarantee the linearity of the velocity. The reaction mixture was transferred to plastic autosampler vials to monitor GPG release. For BMP or LPG monitoring, lipids were extracted as described below and transferred to glass autosampler vials. For kinetic studies, the corresponding standard for GPG, LPG or BMP was measured at increasing concentrations to extrapolate the concentrations of the products. For time-dependent BMP isomer comparison, 100 nM recombinant wild-type PLA2G15-6xHis was mixed with 20 µM BMP substrates in 50 mM sodium acetate:acetic acid at pH 4.5 and 150 mM NaCl. The optimum pH was determined using buffers containing sodium acetate:acetic acid, HEPES and boric acid in their respective pH ranges.
Enzyme activity assays in lysosomal protein extract
Lysosomes obtained from mouse tissues or HEK293T cells were lysed using hypotonic water for 1 h to release the soluble lysosomal proteins. The lysosomal protein extract was then combined with 1 µM 3,3′ S,S BMP and incubated at 37 °C in 50 mM sodium acetate:acetic acid (pH 5) and 150 mM NaCl for the indicated time points. The reaction was stopped using a chloroform:methanol solution in a ratio of 2:1 (v/v) for lipids or 80% methanol for polar metabolite measurements.
Inhibition assays
For inhibition assays, each inhibitor was preincubated with the substrates before either purified PLA2G15-6xHis or lysosomal lysate was added. The reaction buffer contained 50 mM sodium acetate:acetic acid (pH 5.0) and 150 mM NaCl, and we followed the enzyme assays described earlier.
Pulse-chase experiments
A 10× concentration of BMP isomers at a concentration of 100 μM in ethanol, along with fatty acid-free bovine serum albumin (BSA) at the same concentration in PBS, was initially prepared. The concentrated solution was then diluted to a final concentration of 1× in DMEM (Thermo Fisher Scientific). CLN5 PLA2G15 double-knockout cells were seeded into each well of a poly-l-lysine-coated six-well plate. The next day, the medium was gently aspirated, replaced with serum-free DMEM containing 1× solution of BSA-conjugated BMP and incubated for 2 h. After incubation, the cells were washed once with serum-free medium and replaced with complete DMEM-containing vehicle solution (1× PBS), 150 nM wild-type PLA2G15-6XHis or 150 nM PLA2G15(S198A) mutant to chase BMP degradation at varying times. Subsequently, both treated and untreated cells were collected in 200 µl of 80% methanol. Lipids were extracted using a two-phase extraction method involving a 2:1 chloroform:methanol (v/v) mixture, as described below. Finally, lipids were analysed with LC–MS.
Enzyme supplementation
To validate the lysosomal localization of the recombinant enzymes, PLA2G15-6xHis and the mutant were labelled with NHS Alexa 488 dye (Thermo Fisher Scientific) according to the manufacturer’s protocol. Afterwards, 50,000 PLA2G15 knockout or CLN5 PLA2G15 double-knockout cells were seeded with 700-µl DMEM medium on each division of a four-chamber 35-mm glass bottom dish (Cellvis). The cells were then treated with 50 nM fluorescently labelled protein in complete medium for 2 days. After a 2-day incubation, the medium was replaced with 700 µl of DMEM containing 1 µM LysoTracker Red DND-99 (Thermo Fisher Scientific) and 1 µg ml−1 of Hoechst 33342 and then incubated for an extra 30 min. The medium was then replaced with fresh medium and immediately imaged with a Leica LSM 980 ZEN confocal microscope ZEN 3.6 (blue edition). The images were analysed using ImageJ v.2.1.0 and CellProfiler v.4.2.7.
For enzyme supplementation experiments, 600,000 cells were seeded into a six-well plate. After 24 h, the cells were treated with 50 nM PLA2G15 wild-type and mutant proteins in complete medium. After 48 h, lipids were extracted as described above and analysed using lipidomics.
Lipid and polar metabolite extraction
Samples were processed for lipid or polar metabolite extraction, following a previous protocol17. For lipid extraction, 950 µl of 2:1 (v/v) chloroform:methanol containing SPLASH LIPIDOMIX internal standard mix at a concentration of 750 ng ml−1 was added to all the samples. The mixture was then vortexed for 1 h at 4 °C. Subsequently, 200 µl of 0.9% (w/v) NaCl was added, and the mixture was vortexed for 10 min at 4 °C. The mixture was spun at 3,000g for 15 min at 4 °C. The lower chloroform phase, which contained lipids, was collected and dried using a SpeedVac. The dried lipid extracts were reconstituted in 50 µl of 13:6:1 (v/v/v) acetonitrile:isopropyl alcohol:water and vortexed for 10 min at 4 °C in a cold room. The samples were then centrifuged at maximum speed for 10 min at 4 °C, and 45 µl was transferred into glass vials with inserts for LC–MS analysis. For GPG analysis, samples were extracted using 80% methanol in LC–MS grade water containing 500 nM isotope-labelled amino acids as internal standards (Cambridge Isotope Laboratories). The samples were then vortexed for 10 min at 4 °C, centrifuged at 20,627g and transferred into polar vials for LC–MS.
Untargeted lipidomic analysis
Lipidomic analysis of the samples listed in Extended Data Fig. 9h,i and Supplementary Table 8 was performed at the Core Facility Metabolomics of the Amsterdam UMC. In a 2-ml tube, the following amounts of internal standards dissolved in 1:1 (v/v) methanol:chloroform were added to each sample: BMP(14:0/14:0) (0.2 nmol), ceramide-1-phosphate C1P(d18:1/12:0) (0.125 nmol), D7-cholesteryl ester CE(16:0) (2.5 nmol), ceramide Cer(d18:1/12:0) (0.125 nmol), ceramide Cer(d18:1/25:0) (0.125 nmol), cardiolipin CL(14:0/14:0/14:0/14:0) (0.1 nmol), diacylglycerol DG(14:0/14:0) (0.5 nmol), glucosylceramide GlcCer(d18:1/12:0) (0.125 nmol), lactosylceramide LacCer(d18:1/12:0) (0.125 nmol), lysophosphatidic acid LPA(14:0) (0.1 nmol), lysophosphatidylcholine LPC(14:0) (0.5 nmol), lysophosphatidylethanolamine LPE(14:0) (0.1 nmol), lysophosphatidylglycerol LPG(14:0) (0.02 nmol), phosphatidic acid PA(14:0/14:0) (0.5 nmol), phosphatidylcholine PC(14:0/14:0) (2 nmol), phosphatidylethanolamine PE(14:0/14:0) (0.5 nmol), phosphatidylglycerol PG(14:0/14:0) (0.1 nmol), phosphatidylinositol PI(8:0/8:0) (0.5 nmol), phosphatidylserine PS(14:0/14:0) (5 nmol), sphinganine-1-phosphate S1P(d17:0) (0.125 nmol), sphinganine-1-phosphate S1P(d17:1) (0.125 nmol), ceramide phosphocholines SM(d18:1/12:0) (2.125 nmol), sphingosine SPH(d17:0) (0.125 nmol), sphingosine SPH(d17:1) (0.125 nmol) and triacylglycerol TAG(14:0)2 (0.5 nmol). All internal standards were purchased from Avanti Polar Lipids. After addition of the internal standards, 1.5 ml 1:1 (v/v) methanol:chloroform was added before thorough mixing. The samples were then centrifuged for 10 min at 14.000 rpm, and the supernatant was transferred to a glass vial and evaporated under a stream of nitrogen at 60 °C. The residue was dissolved in 150 μl of 1:1 (v/v) methanol:chloroform. Lipids were analysed using an Ultimate 3000 binary HPLC coupled to a Q Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific). For normal phase separation, 5 μl of each sample was injected onto a Phenomenex Luna Silica, 250 × 2 mm, 5 µm 100 Å. The column temperature was held at 25 °C. The mobile phase consisted of 85:15 (v/v) methanol:water containing 0.0125% formic acid and 3.35 mmol l−1 of ammonia (A) and 97:3 (v/v) chloroform:methanol containing 0.0125% formic acid (B). Using a flow rate of 0.3 ml min−1, the liquid chromatography gradient consisted of isocratic at 10% A 0–1 min, ramp to 20% A at 4 min, ramp to 85% A at 12 min, ramp to 100% A at 12.1 min, isocratic at 100% A at 12.1–14 min, ramp to 10% A at 14.1 min and isocratic at 10% A for 14.1–15 min. For reversed phase separation, 5 μl of each sample was injected onto a Waters HSS T3 column (150 × 2.1 mm; 1.8-μm particle size). The column temperature was held at 60 °C. The mobile phase consisted of 4:6 (v/v) methanol:water (A) and 1:9 (v/v) methanol:isopropanol (B), both containing 0.1% formic acid and 10 mmol l−1 of ammonia. Using a flow rate of 0.4 ml min−1, the liquid chromatography gradient consisted of isocratic at 100% A at 0 min, ramp to 80% A at 1 min, ramp to 0% A at 16 min, isocratic at 0% A for 16–20 min, ramp to 100% A at 20.1 min and isocratic at 100% A for 20.1–21 min. Mass spectrometry data were acquired using negative and positive ionization by continuous scanning over the range of m/z 150–2,000 at a resolution of 280,000 full-width at half-maximum. Data were analysed using an in-house developed lipidomics pipeline written in the R programming language (http://www.r-project.org) and MATLAB57. Lipid identification was on the basis of a combination of accurate mass, (relative) retention times, fragmentation spectra (when required), analysis of samples with known metabolic defects and the injection of relevant standards. Lipid classes were defined in our lipidomics pipeline in terms of their generic chemical formula, where R represents the radyl group. Upon import of the lipid database into the annotation pipeline, the generic chemical formula of each lipid class is expanded by replacing the R element with a range of possible radyl group lengths and double bond numbers. The resulting expanded list of chemical formulas was used to calculate the neutral monoisotopic mass of each species. The reported lipid abundances are semiquantitative and calculated by dividing the response of the analyte (area of the peak) by that of the corresponding internal standard multiplied by the concentration of the internal standard (a.u.). The suffix [O] indicates lipids containing an alkyl-ether group, whereas the addition of a quote ‘ indicates an alkenyl-ether group. As no dedicated internal standard for ether lipids is available, we used the lysophosphatidylcholine, phosphatidylcholine, lysophosphatidylethanolamine and phosphatidylethanolamine internal standards to normalize the corresponding ether lipid species, as described above.
Targeted lipidomics and polar metabolomics
Targeted analyses were adapted and performed, as previously described3,7,31. Lipids were separated on an Agilent RRHD Eclipse Plus C18, 2.1 × 100 mm, 1.8u-BC column with an Agilent guard holder (UHPLC Grd; Ecl. Plus C18; 2.1 mm; 1.8 µm), whereas polar metabolites were separated using hydrophilic interaction chromatography using a SeQuant ZIC-pHILIC 50 × 2.1 mm column (MilliporeSigma 1504590001) with a 20 × 2.1 mm (MilliporeSigma 1504380001) guard. Before mass spectrometry, the columns were connected to a 1290 LC system for lipid and metabolite separation. The liquid chromatography system was linked to an Ultivo triple quadrupole (QQQ) mass spectrometer with a liquid chromatography–electrospray ionization probe. External mass calibration was performed weekly using a QQQ standard tuning mix. The column compressor and autosampler were held at 45 °C and 4 °C, respectively. For lipidomics, the mass spectrometer parameters included a capillary voltage of 4.4 kV in positive mode and 5.5 kV in negative mode, and the gas temperature and sheath gas flow were held at 200 °C and 275 °C, respectively. The gas flow and sheath gas flow were 10 and 11 l min−1, respectively, whereas the nebulizer was maintained at 45 psi. The nozzle voltages were maintained at 500 in the positive mode and 1,000 in the negative mode. These conditions were held constant for both ionization mode acquisition. For metabolomics, similar conditions were used, except for the gas temperature and sheath gas flow, which were held at 250 °C and 300 °C, respectively. Additionally, the gas flow and nebulizer pressure were 13 l min−1 and 30 psi.
For lipid measurements, 2- to 4-µl injection volumes were used for each sample for polarity switching. A mobile phase with two distinct components (A and B) was used in the chromatographic process. Mobile phase A was a mixture of acetonitrile:water (2:3 (v/v)), whereas mobile phase B was composed of isopropanol:acetonitrile (9:1 (v/v)), both containing 0.1% formic acid and 10 mM ammonium formate. The elution gradient was carried out over a total of 16 min, with an isocratic elution of 15% B for the first minute, followed by a gradual increase to 70% of B over 3 min and then to 100% of B from 3 to 14 min. Subsequently, this was maintained from 14 to 15 min, after which solvent B was reduced to 15% and maintained for 1 min, followed by an extra 2 min for column re-equilibration. The flow rate was set to 0.400 ml min−1.
For polar metabolite measurements, each sample was injected with a volume of 1–2.5 µl. Mobile phase A was composed of 20 mM ammonium carbonate and 0.1% ammonium hydroxide dissolved in LC–MS grade water, whereas mobile phase B was composed of 100% LC–MS grade acetonitrile. Elution was performed over a 10-min gradient; the B component was linearly decreased from 80% to 20% between 0 and 7 min, rapidly increased from 20% to 80% between 7 and 7.5 min and was held at 80% B from 7.5 to 10 min. The flow rate was set to 0.15 ml min−1.
The QQQ was set to operate in multiple reaction monitoring (MRM) to analyse compounds of interest. Standard lipids, glycerophosphodiesters and amino acids were optimized using the MassHunter Optimizer MRM, a software used for automated method development. For most species, the two most abundant transitions were selected to detect it. The precursor–product ion pairs (m/z) of the compounds used for MRM are listed in Supplementary Table 10.
Polar metabolites and lipids were annotated, and their abundances were measured using the Qualitative MassHunter acquisition software and QQQ quantitative analysis software (Quant My-Way), respectively. The peak areas of all metabolites were integrated using the retention time and MRM method, and the resulting raw abundances were exported to Microsoft Excel for further analysis. The raw abundances of internal controls, endogenous amino acids and highly abundant lipids were examined to ensure the accuracy of the analysis. Raw abundances were normalized to cell numbers using the abundance of endogenous control lipids in the same sample. Quality control samples were consistently measured to ensure optimal instrument performance and linearity. The average of the blank samples was subtracted from all raw abundances. For fold-change calculations, each sample was divided by the average of wild-type or control samples. For total BMP calculations, the sum of all abundances of the measured BMPs was used to generate these values.
Cholesterol measurement
Cholesterol was detected in the left hemibrains and left liver lobes of four animals per group (three samples each from each animal) using a commercial kit (ab65359; Abcam). Left hemibrains and left liver lobes were homogenized in 4 (v/w) PBS using a UPHO bead mill (GENEYE; SOP NEQU200) at 60 Hz for 55 s. Then, 100 µl of the homogenate was transferred to a fresh Eppendorf tube for further processing, and a 5-µl aliquot was used for total protein determination (Pierce BCA assay; Thermo Fisher Scientific; 23225). For extraction, 200 µl of chloroform:isopropanol:NP-40 (7:11:0.1) was added to a 100-µl aliquot, and the sample was vortexed vigorously two times after 5–10 min of incubation phase in between. The extract was then centrifuged for 5–10 min at 15,000g. The entire chloroform phase was transferred to a new tube and air-dried at 50 °C for up to 24 h. The dried lipids were dissolved by vortexing with 100 μl of Assay Buffer II (from kit ab65359) and stored at −80 °C until further use. Cholesterol was assessed in parallel in all samples following the protocol of the commercially available Cholesterol/Cholesteryl Ester Quantitation Assay Kit (ab65359). Fluorescence was measured using a microplate reader (Cytation 5; SOP NEQU201) at excitation/emission = 535/587 nm.
Immunoblotting
Purified proteins were run on SDS–PAGE (Thermo Fisher Scientific) at 120 V and transferred to polyvinylidene difluoride membranes at 40 V for 2 h. The membranes were then blocked with 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween 20 (TBST) for 30 min and incubated overnight with primary antibodies in 5% BSA in TBST at 4 °C using dilutions listed in the antibody table. After incubation, the membranes were washed three times with TBST for 5 min per wash and then incubated with appropriate secondary antibodies diluted 1:3,000 in 5% BSA for 1 h at room temperature. Membranes were then washed three times with TBST followed by ECL2 western blotting substrate (Thermo Fisher Scientific) before visualization.
Thermal stability assay
A Tycho NT.6 instrument (NanoTemper Technologies) was used to determine the melting stability of human wild-type PLA2G15-6xHis and S198A. Purified proteins (5 µM) in 50 mM sodium acetate–acetic acid (pH 5.0) and 150 mM NaCl were loaded into Tycho NT.6 capillaries. The melting temperature (Tm) was obtained by plotting the absorbance ratio of solvent-exposed tryptophan versus buried tryptophan (350 nm/330 nm) over a temperature gradient and determining the inflection temperature from the first derivative.
Microscale thermophoresis
Recombinant human PLA2G15-6xHis S198A was labelled using a RED-NHS Protein Labeling Kit, following the manufacturer’s protocol. A Monolith NT.115 instrument was used for binding experiments. A 100 nM labelled protein was incubated with serial dilutions of substrates in 50 mM sodium acetate–acetic acid (pH 5.0), 150 mM NaCl and 5 µM BSA for 30 min at room temperature. Using premium capillaries, binding measurements were conducted at 20%, 40% or 60% microscale thermophoresis power for 30 s with 5 s of cooling. The dissociation constant Kd was derived by plotting the fraction bound against logarithmic substrate concentrations according to the law of mass action.
Synthesis of GPG
Standard GPG was prepared for kinetic assays. Phosphatidylglycerol (18:1/18:1; Avanti) was saponified to produce GPG according to the following protocol: 100 mg of phosphatidylglycerol was dissolved in a 20-ml scintillation vial that contained a 12-ml solution of 2:1 chloroform:methanol with a magnetic stirring bar. NaOH (2 M; 4 ml) was added to the mixture, and the reaction was stirred for 2 h at room temperature. To neutralize the reaction, 2 M HCl (4 ml) was used, and GPG was extracted with 8-ml water into separate vessels twice. After mixing the aqueous phases, they were frozen in liquid nitrogen, lyophilized and cleaned with 8 ml of chloroform. To separate the crude product from the inorganic salts, lyophilates were dissolved in a minimal amount of methanol. The desired products were then extracted from the precipitate using filter paper.
Thin-layer chromatography
Wild-type PLA2G15-6xHis (100 nM) was mixed with 20 µM BMP and incubated at 37 °C for 1 h, as described for the enzyme assay. Two microlitres of the reaction mixture was spotted on silica and optimized to observe product release. The silica was placed in a thin-layer chromatography solvent (CHCl3/CH3OH/concentrated ammonia/water (90:54:5.5:2 (v/v)) and dipped into an iodine stain (iodine crystals and silica gel mixture) after the solvent run before visualization.
DiffDock docking
The structure of Homo sapiens PLA2G15 was obtained from the AlphaFold entry Q8NCC3 (UniProt). The predicted protein structure was prepared for docking using ChimeraX to add hydrogen according to the expected protonation state at pH 5. The substrates were drawn in ChemDraw to extract the SMILES string. The structure of the protein (Protein Data Bank format) and the SMILES string were used as inputs for DiffDock. DiffDock was run on a web server using the following parameters: 20 inference steps, 18 actual inference steps and 40 samples. The docking results with the best confidence scores were used for analysis using ChimeraX.
siRNA knockdown in mouse BMDMs
A total of 100,000 BMDMs isolated from Gba+/+ or Gba+/D409V mice were seeded into each well on each division of a four-chamber 35-mm glass bottom dish (Cellvis) that contains 750-µl complete DMEM. The next day, the cells were transfected with scramble siRNA (Thermo Fisher Scientific; 4390844) or two siRNAs targeting mouse PLA2G15 (Thermo Fisher Scientific; s101347 and s101348) using RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. Three days after transfection, the cells were subjected to the LysoFQ assay, as described below.
LysoFQ–GCase activity assay
About 50,000 bone marrow-derived macrophages were seeded with 700 µl of macrophage medium on each division of a four-chamber 35-mm glass bottom dish (Cellvis). In the non-siRNA experiment, 300 nM recombinant wild-type or mutant PLA2G15 or vehicle solution (1× PBS) was added during cell seeding. After overnight incubation, 10 µM LysoFQ-GBA probe58 was added to the medium and incubated for 30 min. After incubation, the medium was washed once with the macrophage medium and then replaced with fresh medium containing 100 nM AT3375 (GCase inhibitor) to stop the reaction. The cells were then imaged with a Leica LSM980 Zen confocal microscope ZEN 3.6 (blue edition), and images were analysed using ImageJ v.2.1.0.
Haploid genetic screens
To identify regulators of intracellular cholesterol staining, we prepared libraries of mutagenized Hap1 wild-type and NPC1-deficient cells using a gene-trap retrovirus expressing blue fluorescent protein, as previously described59. HAP1 cells were acquired under licence from the Whitehead Institute and the Netherlands Cancer Institute and confirmed using genetic analysis. For each genotype, the mutagenized cells were cultured and expanded for 1 week after mutagenesis. Subsequently, the cells were washed with PBS, dissociated, pelleted and fixed with BD Fix Buffer I (BD Biosciences) for 10 min at 37 °C. After washing once with fluorescence-activated cell sorting (FACS) buffer (PBS containing 1% BSA) and once with PBS, the cells were pelleted and resuspended in permeabilization buffer (PBS containing 0.1% saponin). The cells were then incubated for 5 min at room temperature while being continuously rotated. The cells were pelleted, washed twice with FACS buffer and incubated with FACS buffer containing 3 µg ml−1 Alexa Fluor 647-labelled PFO60 and 1 µg ml−1 of 4′,6-diamidino-2-phenylindole for 45 min at room temperature while being continuously rotated. The cells were subsequently pelleted and washed twice with FACS buffer before sorting into two populations of cells (PFO-LOW and PFO-HIGH), which represent approximately 5% of the lowest and highest cholesterol-containing cells from the total cell population, respectively. In addition, the cells were sorted in parallel for haploid DNA content (G1 phase) using the 4′,6-diamidino-2-phenylindole signal. Cell sorting was carried out on a FACSAria III Cell Sorter (BD Biosciences) until approximately ten million cells from each population were collected. The sorted cells were pelleted, and genomic DNA was isolated. The gene-trap insertion sites of each sorted cell population were amplified and mapped by deep sequencing. For each gene in a single screen, a mutation index was calculated corresponding to the ratio of the number of disruptive integrations per gene in both populations normalized by the number of total integrations in each channel (that is, PFO-LOW and PFO-HIGH), as previously described59. Genes significantly enriched for disruptive gene-trap integrations in either the PFO-HIGH or PFO-LOW query populations were identified using two-sided Fisher’s exact test. The resulting P values were adjusted for several testing using the Benjamini–Hochberg false discovery rate correction.
siRNA knockdown in fibroblasts derived from patients with NPC1
Two fibroblast cell lines derived from patients with NPC1 (GM03123 and GM18453) were acquired from Coriell with agreement. From these two fibroblast cell lines, 30,000 cells were seeded in each well of an eight-chamber cell culture slide (CELLTREAT) that contains 500-µl complete DMEM. The next day, cells were transfected with scrambled siRNA (Thermo Fisher Scientific; 4390844) or two siRNAs targeting human PLA2G15 (Thermo Fisher Scientific; s24296 and s24297) using RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instruction. Two days after transfection, the medium was gently aspirated, and cells were washed once with 800-µl PBS, followed by immediate 15-min fixation with 200 µl of 4% paraformaldehyde in PBS. The cells were then washed once with 800-µl PBS and stained with 200 µl of 1 mg ml−1 filipin (Cayman). Slides were sealed with glass coverslips and mounting solution (Thermo Fisher Scientific) and imaged using a Leica LSM980 ZEN confocal microscope ZEN 3.6 (blue edition).
Data preparation and statistics
GraphPad Prism v.10.2 was used for quantitative data presentation, and figures were assembled in Adobe Illustrator 2023. All statistical analyses were performed using Prism software. Unless otherwise stated in the legends, all measurements were derived from independent samples or biological replicates. The Western blot data displayed were representative experiments. ChemDraw v.22.2.0 was used in the chemical structures depicted in Figs. 1a–c, 2a and Extended Data Fig. 5h.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.