Mice
Mouse lines and breeding
All experiments followed King’s College London Biological Service Unit’s guidelines and the European Community Council Directive of 24 November 1986 (86/609/EEC). Animal work was carried out under licence from the UK Home Office following the Animals (Scientific Procedures) Act 1986. Similar numbers of male and female mice were used in all experiments. Animals were maintained under standard laboratory conditions on a 12:12 h light:dark cycle in individual ventilated cages at 22 ± 2 °C and 55 ± 10% relative humidity with water and food ad libitum. Sample sizes in animal experiments were chosen to match standards in the field. Experimental conditions were randomized or alternated within litters where possible. The Pvalbcre/+ mice used in most experiments were generated by crossing Pvalbcre/+ mice (B6.129P2-Pvalbtm1(cre)Arbr/J, JAX017320) with CD1 mice (Crl:CD1[ICR], Charles River). To generate Pvalbcre/Flp;RCLChR2/+, Sstcre/+;PvalbFlp/+;RCLChR2/+ and Vipcre/+;PvalbFlp/+;RCLChR2/+ mice, we first generated PvalbFlp/Flp;RCLChR2/ChR2 mice by crossing PvalbFlp/Flp mice (B6.Cg-Pvalbtm4.1(flpo)Hze/J, JAX022730) with RCLChR2/ChR2 (Ai32) mice (B6;129S-Gt(ROSA)26Sortm32(CAG-COP4*H134R/EYFP)Hze/J, JAX012569). These mice were then crossed with Pvalbcre/cre, Sstcre/cre (Ssttm2.1(cre)Zjh/J, JAX013044) or Vipcre/cre (Viptm1(cre)Zjh/J, JAX010908) mice. PvalbFlp/+;NeuroD6cre/+ mice were generated by crossing PvalbFlp/+ mice with Nexcre/+ (Neurod6tm1(cre)Kan) mice70.
Contextual fear conditioning
The day prior to conditioning, animals were briefly handled by the experimenter for 1–2 min. On the following day, mice were transferred to the experimental room in separate cages and allowed to acclimatize for 15–20 min before being placed in the experimental chamber (VFC-008 Fear Conditioning Chamber in a NIR-022SD Sound attenuating Cubicle, Med Associates). Mice received shocks following a standard protocol71. Animals were allowed to acclimatize for 3 min, followed by three mild foot shocks (2 s, 0.7 mA), with 30-s intervals between each shock. Post-shock freezing behaviour was measured for 30 s. The animals were returned to the holding cages and placed back into their home cages 15 min later. For the retrieval experiment, mice were returned to the experimental room in the same holding cages used the previous day. After a 15-min acclimatization period, they were placed in the experimental chamber, and freezing behaviour was recorded for 5 min. Freezing was defined as the absence of movement except for respiration and was measured and analysed using Video Freeze (Med Associated).
Doxycycline treatment and CRAM timeline
For CRAM experiments, mice were put on doxycycline-enriched diet (Envigo, TD.10483, 40 mg kg−1) immediately after surgery and left on this diet for a minimum of 10 days to allow for expression of the construct without expression of tdTomato. Forty-eight hours before cFC, mice were moved back to standard food to allow tdTomato expression following FOS expression upon cFC. In experiments lasting more than 48 h, mice were put back on doxycycline-enriched diet 48 h after cFC to prevent unspecific tdTomato expression.
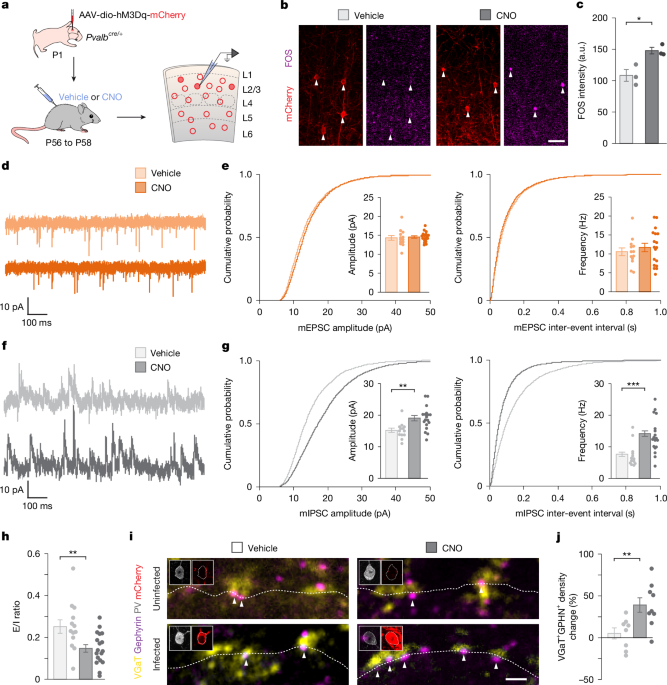
CNO treatment
Mice between P42 and P70 were treated with vehicle or CNO (Tocris, 4936) as indicated. Mice used for FOS experiments received a single injection of vehicle or CNO (1 mg kg−1) and were perfused 2 h later. In all other experiments, mice were treated with vehicle or CNO (1 mg kg−1) for ~48 h via 5 injections. Injections were administered starting 2 days before the experimental day and took place in the morning (between 08:00 and 10:00) and in the evening (between 18:00 and 21:00). A fifth injection was administered between 08:00 and 10:00 on the day of the experiment. CNO was dissolved in 0.5% dimethyl sulfoxide (DMSO, Sigma, D0418) in 0.9% saline and stored at −20 °C.
Histology
Immunohistochemistry
Mice were deeply anaesthetized with pentobarbital before being transcardially perfused with 0.9% saline, followed by 4% paraformaldehyde in PBS. Brains were post-fixed for 2 h in 4% paraformaldehyde at 4 °C, followed by cryoprotection in 15% and 30% sucrose. Brains were sectioned at 40 µm on a sliding microtome (Leica SM2010R) and stored in ethylene glycol (30% ethylene glycol (Merck, 324558), 30% Glycerol (Merck, G5516) in PBS) at −20 °C. Free-floating sections were washed in 0.25% Triton X-100 (Merck) in phosphate-buffered saline (PBS) before 2 h incubation in blocking buffer containing 0.25% Triton X-100, 10% serum and 2% bovine serum albumin in PBS. Sections were incubated overnight at 4 °C in a blocking buffer with primary antibodies. The next day, sections were washed in 0.25% Triton X-100 in PBS and incubated for 2 h in a blocking buffer with secondary antibodies at room temperature. Sections were washed in PBS and stained with 5 µM 4′,6-diamidino-2-phenylindole (DAPI) (Merck) in PBS if required. Sections were dried and mounted with Mowiol/DABCO (8% Mowiol (SLS, 81381), 2% Dabco (Merck, D27802) in PBS). The following primary antibodies and concentrations were used: goat anti-mCherry (1:500, Antibodies-Online, ABIN1440057), dsRed anti-rabbit (1:500, Clontech, 632496), FOS anti-rabbit (1:200, Merck, ABE457), gephyrin anti-mouse-IgG1 (1:500, Synaptic Systems, 147011), VGaT anti-guinea pig (1:500, Synaptic Systems, 131004), parvalbumin anti-chicken (1:250, Synaptic Systems, 195006), parvalbumin anti-mouse (1:3000, Swant, 235) and synaptotagmin-2 anti-mouse-IgG2 (1:250, ZFIN, ZDB-ATB-081002-25). The following secondary antibodies and concentrations were used: Donkey anti-Chicken 405 (1:200, Jackson, 703-475-155), Goat anti-Mouse-IgG1 488 (1:500, Molecular Probes, A21121), Donkey anti-Rabbit 488 (1:400, Thermo Fisher Scientific, A21206), Donkey anti-Rabbit 555 (1:400, Molecular Probes, A31572), Donkey anti-Goat 555 (1:400, Invitrogen, A21432), Goat anti-Mouse-IgG2 647 (1:500, Molecular Probes, A21241), Donkey anti-Guinea Pig 647 (1:250, Jackson, 706-605-148) and Donkey anti-Mouse IgG1 647 (1:400, Thermo Fisher Scientific, A31571).
Single-molecule fluorescence in situ hybridization
Brains were perfused, sectioned and stored in RNase-free solutions. Mice were perfused as described above and post-fixed for 24 h before cryoprotected in 15% and 30% sucrose. The brains were sectioned at 30 µm on a sliding microtome and stored at 20 °C. Sections were mounted on RNase-free SuperFrost Plus slides (Thermo Fisher) and probed following the RNAscope Multiplex Fluorescent Assay v2 protocol (ACDBio 323110). The following probes from ACDBio were used: Mm-Scg2-C1 (477691), Mm-Vgf-C1 (517421) and Mm-Pvalb-C2 (421931).
Image acquisition
Image acquisition and image analysis was performed blind to the treatment condition (CNO or vehicle) of the sample. Images were acquired using an inverted SP8 confocal microscope using the LAS AF software version 3.5.7.23225 (Leica). Different experimental conditions within an experiment were always imaged in parallel, and imaging settings were kept constant for each experiment. All images were taken at 200 Hz acquisition speed and 1,024 × 1,024-pixel resolution. Infection density and FOS intensity were imaged using a 10× objective (0.8800 pixels per µm). RNAscope images were acquired using a 63× objective (5.5440 pixels per µm). Synaptic quantification images were taken using a 100× objective at 1.75× digital zoom (15.4000 pixels per µm). Imaging was restricted to S1 layer 2/3 unless otherwise stated. To image neighbouring cells, we aimed to image an infected and an uninfected cell as close as possible together at the same distance from the pial surface (for example, after imaging an infected PV+ interneuron, an effort was made to image an uninfected PV+ interneuron nearby and at a similar depth from the pial surface).
Image analysis
FOS intensity was analysed using a custom MATLAB script. Cell bodies were segmented using disk morphological shape function, size, and intensity thresholding to create individual regions of interest (ROIs). Layer 2/3 was determined using DAPI counterstaining. ROIs in layer 2/3 were used to measure average signal intensity in the FOS channel.
For single-molecule fluorescence in situ hybridization experiments, the number of mRNA particles was determined using a custom MATLAB script. In brief, for each image, background subtraction was applied. The contour of Pvalb+ cells was then used to draw an ROI and labelled as mCherry+ or mCherry− after visual inspection. Within each ROI, the total area of the signal was measured. This area is then divided by the area of a single mRNA particle (∼0.16 µm2), which estimates the number of mRNA particles in the ROI. We applied intensity correction to normalize signal intensity differences between brains. For this, we measured the average signal intensity of each image, excluding the ROI of the PV+ interneurons and calculated the average signal intensity for each brain. We normalized these values to the lowest value to get the intensity ratio. We then divided the number of mRNA particles by the intensity ratio.
Synaptic densities were analysed using a custom FIJI script, as previously described32. Background subtraction, Gaussian blurring, smoothing and contrast enhancement were applied in all channels. Cell somata were drawn automatically or manually on the basis of intensity levels of parvalbumin staining to create a mask of the soma surface and measure its perimeter. Presynaptic boutons and postsynaptic clusters were detected automatically on the basis of thresholds of intensity. Thresholds for the different synaptic markers were selected from a set of random images before quantification, and the same threshold was applied to all images from the same experiment. The ‘Analyze Particles’ and ‘Adjustable Watershed’ tools were applied to the synaptic channels, and a mask was generated with a minimum particle size of 0.05. The soma mask and the corresponding synaptic masks were merged to quantify the number of puncta contacting the soma and dendrite. Puncta were defined as presynaptic boutons when they were located outside the soma or dendrite and had ≥0.1 µm2 colocalizing with the soma or dendrite perimeter. Puncta were defined as postsynaptic clusters inside a soma or dendrite and had ≥0.2 µm2 colocalising with the soma or dendrite perimeter. Synapses were defined as presynaptic boutons and postsynaptic clusters contacting each other with a colocalization area of ≥0.03 µm2 of their corresponding masks. Classification of cells as uninfected or infected was performed manually on the basis of the absence or presence of mCherry signal, respectively. The density change was calculated as:
$${\rm{Density}}\;{\rm{change}}=\left(\frac{{\rm{Synapse}}\;{\rm{density}}\;{\rm{infected}}\;{\rm{cells}}}{{\rm{Synapse}}\;{\rm{density}}\;{\rm{uninfected}}\;{\rm{cells}}}-1\right)\times 100$$
Viruses
Virus production
AAV8 viruses were produced in HEK293FT cells grown on 5 plates (linear PEI, Polysciences Europe, 23966-100) or 10 plates (branched PEI, Sigma-Aldrich, 408727) of 15 cm diameter until they reached 60% confluency. Cells were grown on DMEM (Gibco 21969-035) + 10% fetal bovine serum (FBS) (Gibco 10500-064) + 1% penicillin/streptomycin (Gibco 15140-122) and 10 mM HEPES. AAVs were produced using polyethylenimine (PEI) transfection of HEK293FT cells with a virus-specific transfer plasmid (70 µg per 10 plates) and a pDP8.ape helper plasmid (300 µg per 10 plates, PF478 from PlasmidFactory). The helper plasmid provided the AAV Rep and Cap functions and the Ad5 genes (VA RNAs, E2A and E4). The DNA and PEI were mixed in a 1:4 ratio in uncomplemented DMEM and left at room temp for 25 mins to form the DNA–PEI complex. The transfection solution was added to each plate and incubated for 72 h at 37 °C in 5% CO2. The transfected cells were then scraped off the plates and pelleted. The cell pellet was lysed in buffer containing 50 mM Tris-Cl, 150 mM NaCl, 2 mM MgCl2, and 0.5% sodium deoxycholate and incubated with 100 U ml−1 benzonase (Sigma, E1014 25KU) for 1 hr to dissociate particles from membranes. The particles were cleared by centrifugation, and the clear supernatant was filtered through 0.8 µm (Merck Millipore Ref SLAA033SS) and 0.45 µm (Merck Millipore Ref SLHA 033SS) filters. The viral suspension was loaded on a discontinuous iodixanol gradient using 4 layers of different iodixanol concentrations72 of 15, 25, 40 and 58% in Quick-seal polyallomer tubes (Beckman Coulter, 342414) and spun in a VTi-50 rotor at 50,000 rpm for 75 min at 12 °C in an Optima L-100 XP Beckman Coulter ultracentrifuge to remove any remaining contaminants. After centrifugation, 5 ml were withdrawn from the 40/58% interface using a G20 needle. The recovered virus fraction was purified by first passing through a 100-kDA molecular mass cut-off centrifugal filter (Sartorius VIVASPIN VS2041) and then through an Amicon Ultra 2 ml Centrifugal filter (Millipore UFC210024). Storage buffer (350 mM NaCl and 5% Sorbitol in PBS) was added to the purified virus, and 5 µl aliquots were stored at −80 °C.
The following viruses were used in this study: AAV8-hSyn-DIO-hM3D(Gq)-mCherry (Addgene 44361), AAV8-hSyn-DIO-mCherry (Addgene 50459), AAV8-hSyn-DIO-hM4D(Gi)-mCherry (Addgene 44362), AAV8-hSyn-fDIO-hM3D(Gq)-mCherry (Addgene 223652), AAV8-hSyn-flx-fDIO-HA-Rpl10a-T2A-Myc-hM3D(Gq) (Addgene 223654), AAV8-hSyn-DIO-hM3D(Gq)-mCherry-shLacZ-CWB (shRNA sequence: AAATCGCTGATTTGTGTAGTC) (Addgene 223660), AAV8-hSyn-DIO-hM3D(Gq)-mCherry-shScg2-CWB (shRNA sequence: GCAGACAAGCACCTTATGAA) (Addgene 223659), AAV8-hSyn-DIO-hM3D(Gq)-mCherry-shVgf-CWB (shRNA sequence: GACGATCGATAGTCTCATTGA) (Addgene 223658), AAV8-hSyn-DIO-Vgf-T2A-mCherry-CW3SL (Addgene 223661), AAV8-hSyn-DIO- mCherry-CW3SL (Addgene 223662), and AAV8-CRAM-tdTomato (Addgene 84468). AAV8-hSyn-DIO-hM3D(Gq)-mCherry, AAV8-hSyn-DIO-mCherry and AAV8-hSyn-DIO-hM4D(Gi)-mCherry were a gift from B. Roth73. pAAV-CWB-EGFP and pAAV-CW3SL-EGFP backbones were used to create plasmids as indicated and were a gift from B.-K. Kaang74. pAAV-hSyn-FLExFRT-mGFP-2A-Synaptophysin-mRuby was used to create fDIO plasmids and was a gift from L. Luo75. pAAV-CRAM-tdTomato was a gift from Y. Lin47. All viruses were diluted to a titre between 5 × 1011 and 8 × 1011 to achieve sparse infection.
Stereotactic injections
Juvenile injection
P1 to P3 pups were anaesthetized with isoflurane for juvenile injections and mounted on a stereotactic frame (Stoelting). Mice used for vTRAP or RNA-seq experiments received six 150 nl injections at a speed of 10 nl s−1 in S1 in the left hemisphere. All other mice received three 150 nl injections at a speed of 10 nl s−1 in S1 in the left hemisphere. For vTRAP experiments, viruses were supplemented with green fluorescent beads (LumaFluor) to label the injection sites. Mice were allowed to recover in a heated recovery chamber (Vettech, HEO11) before returning to the home cage.
Adult injections
P42 to P60 mice were anaesthetized with isoflurane and mounted on a stereotactic frame. A small incision was made in the skin over the injection area on the right hemisphere, and the skull was cleaned using a cotton swab. Two holes were drilled using a rotary drill (Foredom, K.1070) at the following coordinates (from Bregma): (1) anteroposterior +1.0 mm and mediolateral −3.2 mm; and (2) anteroposterior +1.5 mm and mediolateral −3.2 mm for injections in S1, or (1) anteroposterior -2.0 mm and mediolateral −1.7 mm; and (2) anteroposterior −1.5 mm and mediolateral −1.4 mm for injection in hippocampal CA1. Mice received two 300 nl injections at a 100 nl min−1 speed at each location (depth S1: 0.3 mm, depth CA1: 1.2 mm). After injection, the injection capillary was left in place for three minutes before retraction. Following injections, the skin was sutured with absorbable Vicryl sutures (Ethicon, W9500T), and mice were allowed to recover in a heated recovery chamber at 36 °C before being returned to the home cage.
Electrophysiology
Slice preparation
Mice (P42 to P70) were deeply anesthetised with an overdose of sodium pentobarbital and transcardially perfused with 10 ml ice-cold N-methyl-d-glucamine (NMDG) solution containing (in mM) 93 NMDG, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 10 MgSO4 and 0.5 CaCl2 (300–310 mOsm, pH 7.3–7.4) oxygenated with 95% O2 and 5% CO2. Following decapitation, the brain was quickly removed, and the injected hemisphere was glued to a cutting platform before submerging in ice-cold NMDG solution. Coronal slices (300 µm) were cut using a vibratome (VT1200S, Leica) and placed in NMDG solution at 32 °C for 11 min before being transferred to a holding solution containing (in mM) 92 NaCl, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 2 MgSO4 and 2 CaCl2 (300–310 mOsm, pH 7.3–7.4), oxygenated with 95% O2 and 5% CO2, at room temperature for at least 45 min until recording. All salts were purchased from Sigma-Aldrich.
Electrophysiological recordings
Slices were transferred to the recording setup 15 min prior to the recording while being continuously superfused with recording artificial cerebrospinal fluid (ACSF) containing (in mM) 124 NaCl, 1.25 NaH2PO4, 3 KCl, 26 NaHCO3, 10 Glucose, 2 CaCl2, and 1 MgCl2, continuously oxygenated with 95% O2 and 5% CO2 at 32 °C. Pipettes (3–5 MΩ) were made from borosilicate glass capillaries using a PC-10 pipette puller (P10, Narishige) and filled with intracellular solution containing (in mM) 115 CsMeSO3, 20 CsCl, 10 HEPES, 2.5 MgCl2, 4 Na2ATP, 0.4 Na3GTP, 10 sodium-phosphocreatine, 0.6 EGTA (pH 7.2–7.3, 285–295 mOsm). Cells were visualized with an upright microscope (Olympus) and recorded using a Multiclamp 700B amplifier (Molecular Devices). The signal was passed through a Hum Bug Noise Eliminator (Digitimer), sampled at 20 kHz, filtered at 3 kHz using a Digidata 1440 A (Molecular Devices) and recorded using Clampex 10.7 (Molecular Devices). All cells were recorded in S1 layer 2/3. mEPSCs and mIPSCs were recorded in the presence of 1 µM tetrodotoxin (Tocris, 1069) at a holding voltage of −60 mV and +10 mV, respectively. Synaptic currents were not blocked to record mEPSCs and mIPSCs from the same cells. Cells were excluded if the access resistance (Ra) exceeded 25 MΩ or holding current (Ihold) > 200 pA (at holding voltage (Vhold) = −60 mV). For ChR2 eIPSCs, slices were incubated in recording ACSF supplemented with 5 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX, Tocris, 1045) and 100 μM d-(−)-2-amino-5-phosphonopentanoic acid (d-APV, Tocris, 0106) to block glutamatergic inputs. Cells were held at a holding voltage of +10 mV, and after allowing the cell to settle (approximately 1 min), sIPSCs were recorded. Subsequently, whole-cell series resistance was compensated by 70%. eIPSC were evoked with full-field 5 ms LED pulses (power: PV–PV 10% LED, SOM–PV 10% LED, VIP–PV 100% LED, as indicated for the respective experiments. 100% LED power = 75.1 mW mm−2) through a 4× objective (Olympus) with an inter-stimulus interval of 60 s from a pE100 illumination system (CoolLED). Cells were excluded from analysis if the series resistance changed >20%, Ra >25 MΩ or Ihold > 200 pA (at Vhold = −60 mV), and only cells with both sIPSC and eIPSC recordings were included in the data. Cell-attached recordings were performed using recording electrodes filled with recording ACSF in the presence of CNQX and d-APV, loose seals (around 200 MΩ) were formed on cells, and LED stimulus of various intensities was applied via a 4× objective in the same way as in the eIPSC recordings (see above). Paired-pulse ratios were recorded following stimulation in S1 layer 2/3 using an ISO-STIM 01D stimulator (NPI) and a tungsten stimulation electrode (TST33A20KT, WPI) in the presence of CNQX and d-APV at a holding potential of +10 mV. Stimulation strength was set to evoke a ~300 pA response to the first stimulus. Two 1 ms pulses were given with a 50 ms inter-stimulus interval. PPR was calculated as peak 2/peak 1 after correcting for any residual current at the second pulse.
Electrophysiology analysis
Miniature (mEPSC and mIPSC) and spontaneous (sIPSC) recordings were analysed using Mini Analysis (version 6.0.7, Synaptosoft). E/I ratio was calculated as (excitatory current/second)/(inhibitory current/second). Current per second was calculated as (average charge of an event) × frequency. E/I ratios were calculated for each recorded cell, and only cells with mEPSC and mIPSC recordings that passed quality control (see above) were included. eIPSC, cell-attached and paired-pulse ratio recordings were analysed in Clampfit 10.2 (Molecular Devices).
Biochemistry and RNA-seq
RNA isolation by anti-HA pulldown
The infected cortex from P44 to P70 mice, injected with AAV8-hSyn-flx-fDIO-HA-Rpl10a-T2A-Myc-hM3D(Gq) and treated for 48 h with vehicle or CNO, was rapidly dissected in ice-cold RNase-free PBS, using fluorescent beads as a guide to identifying the infected area, and immediately homogenized in ice-cold homogenization buffer (50 mM Tris-HCl pH 7.5, 100 mM KCl, 12 mM MgCl2, 1 mg ml−1 Heparin (Sigma-Aldrich), cOmplete EDTA-free protease inhibitors (Sigma-Aldrich), 200 U ml−1 RNAsin (Promega), 100 µg ml−1 cycloheximide and 1 mM DTT (Sigma-Aldrich)). Tissue from 4 brains was pooled for every sample. Samples were centrifuged at 2,000g for 10 min at 4 °C, and Igepal-CA630 (Sigma-Aldrich) was added to the samples to a final concentration of 1%. Samples were then centrifuged at 13,000g for 10 min at 4 °C, and the supernatant was added on 100 µl of anti-HA magnetic beads (Pierce 88837, previously washed in homogenization buffer) for 3–4 h at 4 °C with gentle rotation. After incubation, beads were washed 3 times in ice-cold washing buffer (300 mM KCl, 1% Igepal-CA630, 50 mM Tris-HCl pH 7.5, 12 mM MgCl2, 1 mM DTT and 100 µg ml−1 cycloheximide) and eluted in 350 µl of RLT Plus buffer from the RNAeasy Plus Micro kit (Qiagen) supplemented with 2-mercaptoethanol (Bio-Rad).
RNA purification, quantification, and quality check
RNA purification of immunoprecipitated RNA was performed using the RNeasy Plus Micro kit (Qiagen) following the manufacturer’s protocol. RNA quality was checked on a Bioanalyzer instrument (Agilent Technologies) using an RNA 6000 Pico Chip. Only RNA samples with RNA integrity number (RIN) values higher than 9 were used for library preparation and sequencing.
Library preparation and Illumina sequencing
Four biological replicates were analysed for each genotype. The Genomic Unit of the Centre for Genomic Regulation (CRG, Barcelona, Spain) performed the library preparation and RNA-seq. The library was prepared using the SMARTer Ultra Low RNAkit and samples were then sequenced paired-end using an Illumina HiSeq 2500 platform to a mean of approximately 60 million mapped reads per sample.
Bioinformatics
High-throughput sequencing data from PV+ interneurons-derived ribosome-associated mRNAs were processed using the community-curated Nextflow (version 21.03.0.edge, build 5518 (3 May 2021 10:52 UTC), available at https://zenodo.org/record/3490660#.Y8AhHXbP2Uk) RNA-seq pipeline76. Specifically, sequencing reads were quality-controlled by FastQC (available at https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and quality-trimmed by Trim Galore (available at https://zenodo.org/record/5127899#.Y8fdOi-l3UI). Mouse GRCm38/mm10 genome annotation was accessed from Illumina’s iGenomes repository (available at https://support.illumina.com/sequencing/sequencing_software/igenome.html) and used as a reference for read alignment by STAR77 and for gene abundance quantification by Salmon78. Gene-level counts data were imported into R using the tximport package79 and analysed by edgeR80 using the estimateGLMRobustDisp model. Genes with less than 10 reads in at least 4 samples were excluded from the analysis81. The >1.5-fold, false discovery rate < 0.05 and transcript per million >1 cut-off were used to identify genes that respond to PV+ interneuron activation (Extended Data Table 1). Activity-dependent genes were ranked using four scoring criteria: fold change, CoV, STRING (https://string-db.org/) and secretome scores. The fold-change score indicates the magnitude of the response and is scaled for a value between 0 and 1. The CoV score represents the degree of reproducibility and was calculated by scaling the sum of the ratio of the standard deviation to the mean from each sample. The STRING score of each gene signifies its co-interaction with other activation-response genes and was computed by scaling the sum of interaction scores obtained from STRING enrichment analysis42. The secretome score takes up a binary value. Genes that are part of the human secretome82 were assigned a value of 1.
Statistical analyses
All statistical analyses were performed using SPSS (Supplementary table 1). No statistical methods were used to predetermine sample sizes. Sample sizes were chosen on the basis of previous publications in the field. Experimental mice from all genotypes or conditions were processed together. Samples were tested for normality using the Shapiro–Wilk normality test. Differences were considered significant when P < 0.05. Data are presented as mean ± s.e.m. Statistical details of experiments are described in figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.