Human samples
Human CD34+ cells and cells from individuals with MDS were maintained in StemSpan Serum-Free Expansion Media (09650, StemCell Technologies) supplemented with 10 ng ml−1 of recombinant human stem cell factor (SCF) (300-07-50UG, PeproTech), recombinant human thrombopoietin (TPO) (300-18-50UG, PeproTech), recombinant human FLT3 ligand (FLT3L) (300-19-50UG, PeproTech), recombinant human IL-3 (200-03-50UG, PeproTech) and recombinant human IL-6 (200-06-50UG, PeproTech), as previously described51. Human CD34+ cells from healthy individuals were obtained from the Yale Cooperative Center of Excellence in Hematology (YCCEH). BM mononuclear cells from individuals with MDS (MDS3328) were obtained with written informed consent and approval of the institutional review board of the University of Cincinnati and Ohio State University and under the IRB-approved study ID 2008-0021. These samples had been obtained within the framework of routine diagnostic BM aspirations after written informed consent in accordance with the Declaration of Helsinki.
Human plasma samples
Human plasma samples were obtained from multiple sources. Plasma from healthy individuals (young (<65 years), n = 5; old (≥65 years), n = 10) and individuals diagnosed with IBD (n = 8) or MDS (n = 9) were obtained from BioIVT. Plasma from individuals with MDS (n = 20) and AML (n = 15) was obtained from Ohio State University. Plasma from healthy individuals (young (<65 years), n = 6; old (≥65 years), n = 7) and individuals with IBD (n = 3) and CHIP (n = 29) were obtained from individuals undergoing elective total hip replacement surgery under the Mechanisms of Age-Related Clonal Haematopoiesis (MARCH) Study (NHS REC: 17/YH/0382) at the Oxford University Hospital, UK. Plasma from individuals with CHIP (n = 30) was obtained from the University of Cincinnati. All of the participants gave written informed consent in accordance with the Declaration of Helsinki. Detailed information is provided in Supplementary Tables 4 and 5.
Cell lines
THP1 cells were purchased from American Type Culture Collection (ATCC). THP1 cells were cultured in RPMI-1640 medium (SH30027.01, HyClone) supplemented with 10% fetal bovine serum (FBS, S11550, Atlanta Biologicals) and 1% penicillin–streptomycin (SV30010, HyClone). HEK293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM, 10-016-CV, Corning Cell Grow) supplemented with 10% FBS and 1% penicillin–streptomycin. THP1-NF-κB-Blue cells (thp-nfkb, Invivogen) were grown in complete THP1 medium with 100 µg ml−1 normocin and 10 µg ml−1 blasticidin added. As mentioned previously52, all cells were cultured at 37 °C and 5% CO2. Analysis of short tandem repeat loci (STR Profiling, ATCC, 135-XV-10) was performed on all cell lines when received and after experimentation was complete. Authentication reports are provided separately. All cell lines were routinely tested and were confirmed to be negative for mycoplasma.
Inhibitors and reagents
Poly(I:C) (4287) was purchased from Tocris Bioscience. IL-1β (200-01B) was purchased from Peprotech. As previously published53, UC-764865 was initially obtained from the University of Cincinnati–Drug Discovery Center’s compound library and then synthesized and purchased from Wuxi AppTec. ADP-heptose (tlrl-adph-l), MRT67307 (inh-mrt) and Ultrapure-LPS (TLRL-PEKLPS) were purchased from Invivogen. GSK8612 (S8872) and ruxolitinib (S1378) were purchased from Selleckchem. N-Des (aminocarbonyl) AZ-TAK1 (ab143773) was purchased from Abcam. PF-06650833 (PZ0327-5MG) was purchased from Sigma-Aldrich. CA-4948 was purchased from ChemExpress. NIK SIM1 (HY-112433), AZD-1480 (HY-10193), itacitinib (HY-16997), tofacitinib (HY-40354), AKT inhibitor VIII (HY-10355) and trametinib (GSK1120212) were purchased from MedChem Express.
Mice
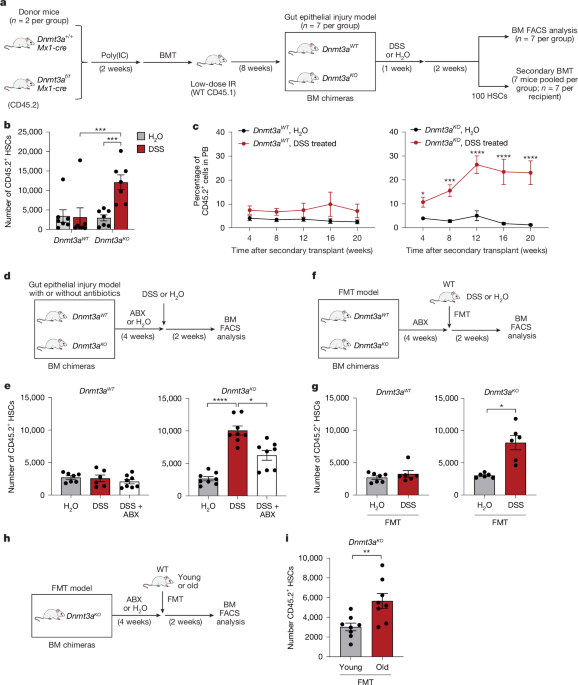
Dnmt3af/f and Mx1-cre+ (obtained from H. L. Grimes laboratory)54, Dnmt3afl-R878H (B6(Cg)-Dnmt3atm1Trow/J, 032289, Jackson Laboratory), Tet2fl/fl (B6;129S-Tet2tm1.1Iaai/J, 017573, Jackson Laboratory), Vav-cre+ (B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J, 008610, Jackson Laboratory), Alpk1−/− (11 bp deletion in exon 3, C57BL/6N-Alpk1em1Fsha/J, 032561, Jackson Laboratory), Tifa−/− (gift from J.-I. Inoue), and UBC-GFP (C57BL/6-Tg(UBC-GFP)30Scha/J, 004353, Jackson Laboratory) mice were maintained on a CD45.2+ C57BL/6 background. cis-NF-κBeGFP reporter mice were provided by C. Jobin55. Throughout the study, CD45.1+ B6.SJL-Ptprca/BoyJ mice were used as recipients for BM transplantation experiments. Littermate controls were used for all experiments. To generate Dnmt3af/fMx1-cre+ mice, Dnmt3af/f and Mx1-cre+ mice were crossed (referred to as Dnmt3a−/− or Dnmt3aKO). To generate Dnmt3a−/−Alpk1−/− mice, Dnmt3a−/− and Alpk1−/− mice were crossed. To generate Dnmt3afl-R878H/+Mx1-cre+ mice, Dnmt3afl-R878H/+ and Mx1-cre+ mice were crossed (referred to as Dnmt3aR878H/+). To generate Tet2f/fVav-cre+ mice, Tet2f/f and Vav-cre+ mice were crossed (referred to as Tet2−/− or Tet2 knockout). To generate Dnmt3a−/− cis-NF-κBeGFP reporter mice, Dnmt3a−/−Mx-cre+ and cis-NF-κBeGFP mice were crossed. All the mice carrying the Mx1-cre allele were given five doses of poly(I:C) every other day at 8–12 weeks of age. Animals of the same age and gender were randomly assigned to experimental groups. Investigators were not blinded.
Husbandry and animal care
All mice were housed in the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility at Cincinnati Children’s Hospital Medical Center, maintained under specific pathogen-free conditions and monitored under tightly controlled settings. They were housed on IVC racks (Allentown Jag 75 Micro-VENT Environmental Systems IVC racks) and kept in individually ventilated polysulfone shoebox cages (Alternative Design), with up to four mice per cage. The cages were supplied with corncob bedding (Bed-o’Cobs 1/4, The Andersons), ad libitum feed (LabDiet, 5010) and enrichment (Twist-n’Rich, The Andersons). All cage components were autoclaved before housing the mice, and cages were changed weekly. Mice had access to ad libitum water through a reverse osmosis autowater system. The mouse room was maintained on an automatic 12 h–12 h light–dark cycle at an ambient temperature of 23 °C and 30–70% humidity, and 5% Clidox-S was used as a disinfectant. Mice were bred, housed and monitored daily by laboratory staff and veterinary personnel to ensure good health, activity and the presence of appropriate food, water and cage conditions. Quarterly testing of pathogens was conducted in sentinel animals housed in the same room. Excluded agents included: Mycoplasma pulmonis, CAR bacillus, Ectromelia, rotavirus (EDIM), Hantaan virus, K virus, lymphocytic choriomeningitis virus, mouse adenoviruses (MAV1, MAV2), mouse cytomegalovirus, mouse hepatitis virus, mouse parvovirus, mouse thymic virus, minute virus of mice, polyoma virus, pneumonia virus of mice, reoviruses (REO3), Sendai virus, Theilers murine encephalomyelitis virus (TMEV), Encephalitozoon cuniculi, Aspiculuris tetraptera, Fur mites (Myocoptes, Radfordia/Myobia) and Pinworms (Aspiculuris tetraptera, Syphacia muris, Syphacia obvelata). All laboratory staff wore personal protective clothing, and all animal procedures were performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital (IACUC) (IACUC2019-0072). Procedures such as blood collection, faecal sample collection and oral gavage were conducted in a biosafety cabinet (NuAire) within the same experimental housing room (6445), eliminating the need to transport mouse cages through the halls during experiments. Throughout the study, care was taken to collect all biological specimens from the mice by a single scientist (K.H.) and processed by another scientist (P.A.) using the same laboratory equipment and reagent kits from the same commercial vendor.
DSS and antibiotics treatment
Mice were treated with 2.5% DSS (w/v) (molecular mass: 36,000–50,000 Da, 216011090, MP Biomedicals) in autoclaved drinking water to induce gut injury-associated colitis, as described previously56. Control mice were time and anatomical location matched and received water only. Mice were monitored daily for weight loss, stool consistency and the presence of frank blood in the stool. Daily assessment of mortality/morbidity was performed, and mice were euthanized if they were in obvious distress (defined as immobility, weight loss >20% or severe bloody diarrhoea), and were therefore not included in the study. Study animals were allowed to recover on regular water for an additional 1–8 weeks. Blood was collected through the submandibular vein and faecal pellets, distal colons and BM were collected for histological analysis and flow cytometry. In parallel experiments, mice were pretreated with a broad-spectrum antibiotics cocktail to deplete endogenous host microbiota as previously described57,58. In brief, in the first week (Monday–Friday), mice received a daily oral gavage with 100 μl of antibiotics cocktail containing kanamycin (4 mg ml−1, Sigma-Aldrich, 60615), gentamicin (0.35 mg ml−1, Sigma-Aldrich, G1914), colistin (0.5 mg ml−1, Sigma-Aldrich, C4461), metronidazole (2.15 mg ml−1, Sigma-Aldrich, M3761) and vancomycin (0.45 mg ml−1, Sigma-Aldrich, V2002). For the next 3 weeks, antibiotics were administered in non-acidified autoclaved water at 0.2 mg ml−1 except for vancomycin, which was maintained at 0.5 mg ml−1. In few experiments wherever noted, in the first week (Monday–Friday), mice received a daily oral gavage of 100 μl of antibiotics containing either vancomycin (1 mg ml−1) to deplete Gram-positive bacteria or cocktail of metronidazole (2.15 mg ml−1), gentamicin (1 mg ml−1) and neomycin (1 mg ml−1) (MGN) to deplete Gram-negative bacteria. For the next 3 weeks, the antibiotics were administered in non-acidified autoclaved water maintained at 1 mg ml−1. Antibiotics water was prepared fresh and replaced weekly to supply fresh antibiotics.
BM transplantation
To model pre-leukaemic clonal haematopoiesis, we generated chimeric mice as follows. In brief, a mixture of 1 × 106 whole BM cells (WBM) was obtained from poly(I:C)-treated wild-type (Dnmt3a+/+Mx1-cre+, called Dnmt3a wild-type) or mutant mice (Dnmt3af/fMx1-cre+, called Dnmt3aKO) or double-mutant mice (Dnmt3af/fMx1-cre+Alpk1KO, called Dnmt3aKOAlpk1KO) (CD45.2+), and transplanted into low-dose (2.5 Gy) irradiated recipient mice (CD45.1+; 6–10 weeks of age). Then, 8 weeks after transplant, chimeric mice were treated with either water or DSS (2.5%) for 1 week, and allowed to recover for 1 more week on water after which flow cytometry was performed on the BM. In a separate experiment, chimeric mice were pretreated with broad-spectrum antibiotics for 4 weeks, and then subjected to DSS for 1 week after which flow cytometry performed on BM. In a separate set of experiments, chimeric mice were treated with either water or ADP-heptose (0.5 mg per kg) through oral gavage for 2 weeks, and flow cytometry analysis of the BM was performed after 2 more weeks. In all of the experiments, secondary transplantation was performed by purifying donor HSCs (CD45.2+Lin−KIT+SCA1+CD150+CD48−) and transplanting 100 HSCs with 200,000 helper WBM cells (CD45.1+) into lethally irradiated (8 Gy) recipient mice (CD45.1+), and donor chimerism in PB examined by flow cytometry.
Quantification of bacterial DNA using qPCR
We used the previously described protocol to examine bacterial translocation into blood59,60. Whole blood was collected by cheek bleeding in sterile BD Microtainer Capillary Blood Collector and Microgard Closure tubes (13-680-62, Thermo Fisher Scientific) on ice from each mouse using Goldenrod Animal Lancets 4 mm (NC9922361, Braintree Scientific), and genomic DNA was extracted using the DNeasy Blood & Tissue Kit (69504, Qiagen). Quantitative PCR (qPCR) was performed using the Femto Bacterial DNA Quantification Kit (E2006, Zymo Research) according to the manufacturers’ instructions. Samples with a Ct value of more than 35 cycles or undetectable were counted as 0 pg ml−1.
In vitro competition assay
The polyvinyl alcohol-based in vitro HSC expansion protocol was adapted as previously described61,62. 50 HSCs from wild-type GFP (C57BL/6-Tg(UBC-GFP)30Scha/J, 004353, Jackson Labs) and 50 HSCs from Dnmt3a−/− mice were sorted directly into each well of a fibronectin-coated 96-well plate (08-774-60, Thermo Fisher Scientific) with Ham’s F12 nutrient mix medium (11765054, Thermo Fisher Scientific) containing final concentrations of 1× penicillin–streptomycin–glutamine (10378-016, Thermo Fisher Scientific), 10 mM HEPES (15630080, Thermo Fisher Scientific), 1× insulin–transferrin–selenium–ethanolamine (ITS-X, 51500056, Thermo Fisher Scientific), 100 ng ml−1 recombinant murine TPO (AF-315-14, Peprotech), 10 ng ml−1 recombinant murine SCF (250-03, Peprotech) and 1 mg ml−1 poly(vinyl alcohol) (P8136, Millipore Sigma) at 1:1 ratio at 37 °C and 5% CO2. Then, 1 µg ml−1 ADP-heptose treatment was started at day 8 after starting the culture when the second medium change was performed and added every 3 days with subsequent medium changes. After 14 days of ADP-heptose treatment, cells were collected, counted using the trypan blue exclusion assay and analysed by flow cytometry. To enumerate cells, a defined number of CountBright Absolute Counting Beads (Thermo Fisher Scientific, C36950) was added to each sample and cell count was back calculated to the proportion of the total that was run through the cytometer.
Immunoblotting
For immunoblots, total protein lysates were obtained from cells by lysing the samples in cold RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% SDS, in the presence of phenylmethylsulfonyl fluoride, sodium orthovanadate and protease and phosphatase inhibitors, as previously described63. After being resuspended in RIPA, cells were lysed by vortex followed by incubation on ice for 20 min. Protein concentration was evaluated using the bicinchoninic acid assay (Pierce, 23225). SDS sample buffer was added to the lysates and the proteins were separated by SDS–PAGE, transferred to PVDF or nitrocellulose membranes (Bio-Rad, 1620112) and analysed by immunoblotting. Western blot analysis was performed using the following antibodies: UBE2N (Abcam, ab25885; Cell Signaling, 6999 or 4919S, 1:1,000), vinculin (Cell Signaling, 13901T, 1:1,000), GAPDH (Cell Signaling, 5174T; D16H11, 1:1,000) phospho-IKKα/β (Ser176/180) (Cell Signaling, 2697, 1:1,000), MyD88 (Cell Signaling, 4283, 1:1,000), TRAF6 (Santa Cruz, sc-7221, 1:1,000), p65 (Cell Signaling, 8242, 1:1,000), phosphor-p65 (Ser536) (Cell Signaling, 3033, 1:1,000), IRAK4 (Cell Signaling, 4363, 1:1,000), IRAK1 (Santa Cruz, sc-5288, 1:1,000), phospho-SAPK/JNK (Thr183/Tyr185) (Cell Signaling, 4668, 1:1,000), SAPK/JNK (56G8) (Cell Signaling, 9258, 1:1,000), phospho-p38 MAPK (Thr180/Tyr182) (Cell Signaling, 4631, 1:1,000), p38 MAPK (Cell Signaling, 9212, 1:1,000), phospho-p44/42 MAPK (ERK1/2, Thr202/Tyr204) (Cell Signaling, 4377, 1:1,000), p44/42 MAPK (ERK1/2) (137F5) (Cell Signaling, 4695, 1:1,000), total IKKα/β (Cell Signaling, 2697, 1:1,000), ALPK1 (Abcam, ab236626), TIFA (Cell Signaling, 61358S, 1:1,000) and actin (Cell Signaling Technology, 4968, 1:1,000), and peroxidase-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 111-035-003, 1:10,000), and peroxidase-conjugated AffiniPure goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, 115-035-003, 1:10,000). The membranes were visualized using ECL Western Blotting Substrate (Pierce, 32106) or SuperSignal West Femto Substrate (Thermo Fisher Scientific, 34096), imaged on the Bio-Rad ChemiDoc Touch Imaging system and analysed using Image lab software v.6.0.1 (Bio-Rad) or Image J (22930834).
Quantitative analysis of TIFAsomes and ADP-heptose in biological samples using multispectral imaging flow cytometry (TIFAsome assay)
THP1 (THP1 TIFA-TdTomato) or THP1 ALPK1KO cells (ALPK1KO-TIFA-TdTomato THP1) (1 × 106) were stimulated with various human plasma samples (100 µl) for 30 min in a 37 °C water bath in a final volume of 200 µl. Cells were collected, washed with PBS + 2% FBS + 2 mM EDTA (MACS buffer) and fixed with 4% paraformaldehyde (15710, Electron Microscopy Sciences). After fixation, cells were washed again and then resuspended in 50 µl MACS buffer. Cells were then analysed for TIFAsome formation on the Amnis Imagestream Mk II Imaging Flow Cytometer ISX-100 (Luminex) according to the manufacturer’s instructions. Downstream analysis was performed using IDEAS analysis software (Amnis). TIFAsome-positive cells were identified by gating on the mean pixel intensity and maximum pixel intensity for bright puncta analysis using the IDEAS Image Data Exploration and Analysis Software. A standard curve was prepared by calculating the percentage of TIFAsome-positive cells using samples that were stimulated with serial increasing doses of ADP-heptose covering the concentration range of 10 to 100,000 ng ml−1. Using the data from the standard curve, the ADP-heptose concentration was extrapolated and estimated in unknown human biological samples using the following calculation: a standard curve was run, and the trend line was created. From the trendline equation y = mx + b, the concentration of ADP-heptose (x) was calculated by x = ((y − b)/m) × d where m is the slope of the trend line, b is the y-intercept, x is ADP-heptose concentration, y is percentage of positive TIFAsome cells and d is the dilution factor of the plasma.
To assess TIFAsome formation in mouse cells, HSPCs were purified from the BM of wild-type and knockout Dnmt3a mice using the CD117 MicroBeads, mouse (Miltenyi Biotech, 130-091-224) and cultured overnight in polyvinyl alcohol (PVA)-based medium on retronectin-coated plates. Cells were then transduced with pCDH-TIFA-TdTomato-GFP lentiviral particles and polybrene (0.8 µl ml−1) using the ultracentrifugation method. In brief, cells were centrifuged at 32 °C and 800g for 1.5 h and then cultured in fresh PVA medium for 3 days, after which KIT+GFP+TdTomato+ cells were sorted. Next, cells were stimulated with ADP-heptose (1 µg ml−1) and collected at serial timepoints (0 h, 0.5 h, 4 h, 24 h, 48 h), and subjected to TIFAsome assay as mentioned above.
Immunofluorescence
TIFA-TdTomato-GFP THP1 cells were suspended at 1 × 106 cells per ml and treated with either human plasma samples (50 µl) in final volume of 200 µl for 30 min or stimulated with ADP-heptose for 30 min. Cells were then washed and spun onto slides using a cytospin at 500 rpm at low acceleration. Slides were then fixed in PBS containing 4% paraformaldehyde and 0.1% Triton X-100. Slides were then blocked for non-specific binding in PBS with 3% bovine serum albumin and 0.1% Tween-20. The slides were mounted with ProLong Gold Antifade Mounting medium. Images were acquired using the Nikon Ni-E Upright widefield fluorescent scope and analysed using Nikon Elements.
In vivo FITC–dextran permeability assay
As previously described28,64, the FITC assay is a measure of total intestinal permeability. In brief, mice were fasted for 5 h before the test at the beginning of the light cycle (12 h cycle) to minimize discomfort. When fasting, mice were transferred to a new cage (to limit coprophagy) without food or bedding but were kept with water bottles in the cage to avoid dehydration. After fasting, blood was collected by cheek bleeding on ice. Immediately after blood collection, the mice were then gavaged with freshly prepared 150 μl of 80 mg ml−1 fluorescently labelled smal-molecule FITC–dextran (4 kDa) (Sigma-Aldrich, 46944-500MG-F) diluted in sterile 1× PBS. Blood collection was repeated at 4 h after gavage and the mice were then returned immediately to their regular cages with bedding, food and water. Plasma was prepared by centrifugation of blood samples at 2,000g for 10 min at 4 °C and protected from light at all times. The FITC–dextran concentration in the plasma was measured using a fluorescence spectrophotometer with emission and excitation wavelengths of 520 nm and 490 nm, respectively.
Haematological and histological analysis
Blood counts were measured using a Genesis blood analyzer (Oxford Scientific). Spleens, femurs and livers were fixed with 10% formalin, sectioned and stained with haematoxylin and eosin (H&E). Distal colonic tissues were fixed in 10% formalin, paraffin embedded and processed for H&E staining as previously described65. DSS-associated experimental colitis severity was assessed in a blinded manner by a pathologist using an established semi-quantitative multiparameter histopathological scoring system based on the following criteria: percentage area involved (0–4), oedema (0–3), ulceration (0–4), crypt loss (0–4) and leukocyte infiltration (0–3).
Flow cytometry and cell sorting
Mice were euthanized using CO2 followed by cervical dislocation. PB was collected into EDTA-coated tubes (22030403, Thermo Fisher Scientific), and hind limb bones (femurs, and tibias) were obtained immediately after euthanasia and stored in cold FACS buffer (1% FBS in DPBS) under sterile conditions. Bones were crushed using a mortar and pestle and then passed through a 40 μm cell strainer (542040, Greiner Bio-one) for various applications. PB and BM cells were labelled with respective antibodies and analysed on a BD LSRII and BD Fortessa X-20 flow cytometers (BD Biosciences), and FACSDiva 8.0 and FlowJo software. For immunophenotypic analysis of PB samples, cells were first lysed with 1× red blood cell lysis buffer (555899, Thermo Fisher Scientific), and then incubated with CD19-PE (115507, BioLegend, 1:100), CD3-PerCpCy5.5 (100218, BioLegend, 1:100), Gr-1-APC (17-5931-81, eBioscience, 1:100) and CD11b-PE Cy5 (15-0112-82, eBioscience, 1:100). For HSPC analysis, BM cells were washed and incubated for 30 min with biotin-conjugated lineage markers (CD11b, Gr1, Ter119, CD3, B220, mouse haematopoietic lineage biotin panel (88–7774-75 eBioscience, 1:50)), followed by staining with streptavidin eFluor450 (48-4317-82, Thermo Fisher Scientific, 1:100), SCA1-PE (12–5981-82, eBioscience, 1:100), KIT-APC Cy7 (135135, BioLegend, 1:100), CD150-PerCp Cy5.5 (115922, BioLegend, 1:100) and CD48-APC (103412, BioLegend, 1:100). HSCs were identified on the basis of the expression of Lin−SCA1+KIT+CD150+CD48−. During in vitro HSC competition assay, to calculate the absolute number of cells whenever required, CountBright Absolute Counting Beads (C36950; Thermo Fisher Scientific, 1:10) were mixed with the cell sample (per well) and assayed by flow cytometry. By comparing the ratio of bead events to cell events, the absolute numbers of cells in the sample were calculated. For all of the experiments involving transplantations, to distinguish donor from recipient haematopoietic cells, PB and BM cells were also stained with CD45.1-Brilliant Violet 510 (110741, BioLegend, 1:100), and CD45.2-FITC (553772, Fisher Scientific, 1:100) or CD45.2-eFluor450 (48–0454-82, eBioscience, 1:100). HSCs were sorted as described previously66. BM cells were first enriched for stem/progenitor cells by using either lineage depletion (mouse total lineage kit, 130-110-470, Miltenyi Biotec, 1:50) or KIT enrichment kit (mouse CD117 microbeads, 130-091-224, Miltenyi Biotec, 1:50). KIT-enriched cells were immunostained for HSPC markers as mentioned above and sorted on the BD FACSAria II sorter (BD Biosciences).
Measurement of cytokines and chemokines by multiplex ELISA
On day 1, wild-type, Dnmt3a−/− and Dnmt3a−/−Alpk1−/− mice were treated with ADP-heptose (0.5 mg per kg) twice (5 h apart). On day 2, mice were treated with ADP-heptose in the morning and the bones collected 5 h later. For preparing BM fluid, 2 femurs from each mouse were sectioned at the two ends and flushed with 200 µl of ice cold PBS containing 1× protease inhibitor (11836153001, Millipore Sigma), and transferred to cold Eppendorf tubes. After centrifugation at 1,000g for 5 min at 4 °C, the supernatant was immediately transferred to ice-cold Eppendorf tubes and frozen at −70 °C until further use. The samples were thawed on ice, vortexed thoroughly before being diluted 1:1 in assay buffer using the mouse cytokine/chemokine magnetic bead panel kit to quantify 32-plex mouse panel (MCYTOMAG-70K; Millipore Sigma).
Colony forming cell assay
Clonogenic progenitor frequency was determined by plating freshly purified mouse LSK cells (5,000 cells per ml) in MethoCult GF M3434 (Stem Cell Technologies) or human patient samples (1,000 CD34+ cells per ml) in MethoCult H4434 (Stem Cell Technologies) in SmartDish meniscus-free 6-well plates in the presence of ADP-heptose (1 µg ml−1) with or without UBE2Ni (10 µM). Cells were incubated at 37 °C and 5% CO2. Colonies were scored at 14 days after plating using STEMVision (StemCell Technologies).
Cell cycle analysis
As described previously67, mice were injected intraperitoneally with EdU (Invitrogen, 1 mg per mouse) and euthanized 6 h later and the EdU incorporation was analysed using the Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit (C10633, Thermo Fisher Scientific).
qPCR with reverse transcription
Total RNA was extracted and purified using the Quick-RNA MiniPrep (Zymo Research, R1055) or RNeasy Micro (Qiagen) kit, and reverse transcription was carried out using the Superscript complementary DNA Synthesis Kit (Invitrogen) or High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR was performed using the Taqman Master Mix (Life Technologies) for mouse Alpk1 (Mm01320377_m1), Tifa (Mm07300088_m1) and Gapdh (Mm99999915_g1).
Plasmids and viral transduction
As described elsewhere34, for generating N-terminal TIFA C-terminally fused to TdTomato, TIFA from human TIFA tagged ORF clone (NM_052864, RC204357, Origene) was amplified and cloned into pTdTomato N1 (54642, Addgene). For expressing TIFA-TdTomato fusion protein in THP1 cells, we cloned the corresponding TIFA-TdTomato cDNA into the pCDH-EF1-MCS-IRES-GFP vector plasmid from Systems Biosciences (CD530A-2). TIFA-TdTomato and pCDH vector were both incubated separately with EcoRI and NotI (New England Biosciences). Insert and vector fragments were run on a 1% agarose gel and extracted from the gel using a Qiagen Gel Extraction Kit (Qiagen). After gel extraction, insert and vector fragments were ligated together using T4 DNA ligase. DH5a competent cells were transformed with ligation products and streaked onto LB agar plates containing ampicillin for selection against negative clones. Colonies were picked, sequenced and a single correct clone was chosen for further study. Viral particles were produced using Mirus Trans-IT LT1 transfection reagent according to manufacturer protocols (Mirus). HEK293T cells were seeded to a confluency of approximately 80%. pCDH-TIFA-TdT-GFP plasmid was incubated with viral packaging plasmids containing gag-pol and VSV-G in the Trans-IT LT1 reagent (MIR 2306, Mirus). Packaged plasmid DNA was then added dropwise to seeded HEK293T. Cells were incubated for 48 h to allow for viral production. The viral supernatant was filtered and added to THP1 cells for transduction. Transduced cells were sorted for GFP+TdTomato+ using the BD FACSAria cell sorter, after which cells were grown in culture.
Generation of mutant cells using CRISPR–Cas9 technology
THP1 IRAK1-knockout, THP1 IRAK4-knockout, THP1 IRAK1/IRAK4-double knockout, THP1 MYD88-knockout and THP1 TRAF6-knockout cells have been described previously68 THP1 TIFA-knockout cells were generated using a modified synthetic gRNA targeting exon 2 of the TIFA gene (Synthego)3 (sgRNA sequence, CAGAUGACGGUUUACCAUCC). THP1 ALPK1-knockout cells were generated using a synthetic multi-sgRNA kit targeting exon 5 of the ALPK1 gene (Synthego gene knockout kit v2; human ALPK1). The sgRNAs used were as follows: sgRNA1, CAUCCUCGCUCGGGACUGUG; sgRNA 2, CUGUAUGGGCUCGACGUCUC; sgRNA3, AGUUCACGGAGAUUCGGGCU. Cells were generated by suspending the parental THP1 cells in buffer R with Cas9-NLS and sgRNA, and electroporated (1,700 mV × 20 ms × 1 pulse) using the Neon Transfection system (Invitrogen). As a control, THP1 PTPRC-knockout (CD45-knockout) cells were also generated using sgRNA targeting exon 2 of the PTPRC gene. The transfected cells were recovered for 48 h in antibiotic-free medium. CD45 deletion was assessed by flow cytometry 5 days after transfection. Deletion for all other proteins was then confirmed by immunoblotting.
In vivo pharmacokinetic analysis
Pharmacokinetic study of synthetic ADP-heptose was performed using our previously established protocol53 in C57BL/6 mice (18 to 22 g). ADP-heptose was administered through oral gavage (0.5 mg per kg per mouse). Blood samples were then collected at 4 h, 8 h, 16 h and 24 h after dosing. Samples from four animals were collected at each timepoint. About 200 µl of blood was collected through the orbital vein from each mouse, processed and analysed by LC–MS. A standard curve was prepared in blood covering the concentration range of 50 to 20,000 pg ml−1. Using the data from the standard curve, calibration curves were generated for pharmacokinetic tests.
Bacterial culture of mouse tissues and human plasma
All of the mouse tissues were collected under sterile conditions with autoclaved tools by the same researcher throughout the study. In brief, PB was collected by cheek bleeding (after sterilizing the cheeks with 70% ethanol wipes) in sterile BD Microtainer Capillary Blood Collector and Microgard Closure tubes on ice from each mouse using Goldenrod Animal Lancets 4 mm. Plasma was then prepared by centrifuging at 5,000 rpm for 10 min. Bones (2 femurs and 2 tibiae per mouse) were grinded using a sterile mortar and pestle using sterile PBS. After red cell lysis using RBC lysis buffer (555899, BD Biosciences), the samples were resuspended in 300 µl of sterile filtered (0.22 μm) PBS + 0.1% l-cysteine (168149, Sigma-Aldrich) and homogenates prepared by homogenizing the samples in hard tissue homogenizing CK28 tubes (P000911-LYSK0-A, Bertin Instruments) using the Minilys homogenizer (Bertin Technologies). One fresh faecal pellet was collected from each mouse and resuspended in 1 ml of sterile PBS + 0.1% l-cysteine and homogenized in soil grinding SK38 tubes (P000915-LYSK0-A, Bertin Instruments) using the Minilys homogenizer. Then, 100 µl of mouse or human plasma, 100 µl of mouse BM homogenate and 100 µl of mouse faecal homogenates were then used for culturing on parafilm sealed Teknova brain heart infusion (BHI) agar plates (50-841-098, Thermo Fisher Scientific) and incubated upside down for 48 h. For all of the experiments, negative-control BHI plates were setup using 100 µl of sterile PBS + 0.1% l-cysteine; no colonies were observed in the negative control plates. Using sterile pipet tip, all of the colonies grown on BHI plates were scraped and mixed grown in 6 ml of liquid BHI broth for 24 h. Later, 750 µl of bacterial suspension was collected for immediate purification of bacterial DNA and remaining suspension used for preparation of lysates.
RNA-seq analysis
Mouse LSK cells were purified from wild-type, Dnmt3a−/− and Dnmt3a−/−Alpk1−/− mice by flow cytometry and treated with ADP-heptose for 90 min in vitro. Total RNA was then extracted using the RNeasy Plus Micro Kit (Qiagen). The initial amplification step for all of the samples was performed using the NuGEN Ovation RNA-Seq System v.2. The assay was used to amplify RNA samples to create double-stranded cDNA. The concentrations were measured using the Qubit dsDNA BR assay. RNA libraries were then created for all samples using the Illumina protocol (Nextera XT DNA Sample Preparation Kit). The concentrations were measured using the Qubit dsDNA HS assay. The size of the libraries for each sample was measured using the Agilent HS DNA chip. The concentration of the pool was optimized to acquire at least 30–40 million reads per sample. The sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. Paired-end FASTQ files were aligned to mm10 (mouse) genomes using HISAT2 (http://www.ccb.jhu.edu/software/hisat) or Tophat (https://ccb.jhu.edu/software/tophat). The feature Counts program (http://subread.sourceforge.net/)69 was used to generate counts for each gene based on how many aligned reads overlap its exons. These counts were then normalized and used to test for differential expression using negative binomial generalized linear models implemented by the DESeq2 R package (v.1.30.1). Further downstream analysis was performed with iGeak software70. We considered a gene as differentially expressed if statistically supported at FDR-adjusted q < 0.1 and a |log2[fold change]| > 0.5. Functional enrichment analysis was performed using the gene set enrichment analysis method71. All RNA-seq data generated in this study are available at the GEO (GSE232794).
Xenograft and in vivo drug treatment
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG)72 mice were bred and maintained by the CCHMC Comprehensive Mouse Core. For patient-derived xenografts, NSG mice (sublethally conditioned with 2 Gy of whole-body irradiation) were injected into the tail vein with healthy CD34+ cells (1 × 106 cells per mouse) and cells from individuals with MDS (5 × 106 cells per mouse) in 200 µl of sterile PBS. Mice were then given sterile water or ADP-heptose (0.5 mg per kg) dissolved in sterile water at the indicated times. Mice were monitored for human engraftment in BM aspirates. In brief, 1 × 106 BM cells from each sample were incubated with anti-human CD45 (555485, BD Biosciences, 1:100) and anti-human CD33 (555450, BD Biosciences, 1:100) antibodies in a solution of PBS, 0.2% FBS for 30 min on ice. Cells were washed once with PBS, resuspended in PBS with 0.2% FBS and immediately analysed by flow cytometry.
NF-κB activation reporter
THP1-Blue NF-κB SEAP reporter cells (thp-nkfb, Invivogen) were grown at 20,000 cells per well (200 µl) in a 96-well plate with the indicated agonists and inhibitors for 24 h. The next day, QuantiBlue Reagent (Invivogen, rep-qbs2) was warmed to 37 °C in a water bath and 180 µl was added to each well of a new, clean 96-well plate. The incubated cells were centrifuged and 20 µl of supernatant from each well was pipetted into the respective 180 µl QuantiBlue Reagent well, in triplicate. The reaction was mixed and incubated for 1 h, when a colour gradient could be seen. The absorbance was read at 630 nm for a final readout. For analysis, the medium absorbance was subtracted, experimental values were normalized to the vehicle control and triplicates were averaged.
Bacterial DNA extraction for 16S rRNA-seq
To examine the gut microbiota diversity and phylogenetics analysis, 16S rRNA-sequencing (rRNA-seq) was performed on faecal DNA isolated in a clean room environment from fresh faecal pellets using a previously described protocol73. In brief, faecal pellets were collected using sterile pipet tips in the biosafety cabinet from each mouse at the same time period of the day by same mouse handler throughout the study in sterile soil grinding SK38 tubes on ice. DNA was immediately extracted using the ultraclean QIAamp Fast DNA Stool Mini Kit (51604, Qiagen) according to the manufacturer’s recommendations using mechanical bead beating and a chemical-lysis-based approach to minimize kit contamination (kitome) and maintain low microbial biomass74. All of the mice were always housed in the same room. In a separate set of experiments, DNA was extracted from the bacterial suspension of various mouse tissues and human plasma using the same protocol. Ultraclean reagents were always used and all of the pre- and post-PCR and sequencing experiments were performed in separate designated area.
Preparation of bacterial lysates for mass spectrometry
To measure ADP-heptose, lysates were prepared as described previously44. In brief, bacterial culture suspensions of various mouse tissues and human plasma samples were first centrifuged at 4,000g for 5 min. Next, the pellets were suspended in 1 ml of sterile water. Bacterial cells were then lysed by heating at 95 °C for 15 min. Next, the lysates were centrifuged at 4,000g for 3 min and the supernatants were filtered through a 0.20 µm syringe filter using 1 ml syringe and needle. Finally, lysates were stored at −80 °C until further processing for MS. According to a previously established protocol, the lysates were prepared4. In brief, 500 µl of the cleared bacterial lysate was thawed from −80 °C and extracted by addition of 1 ml chloroform:methanol (v/v, 2:1) and vortexed and centrifuged (5 min, 10,000g). The aqueous phase was then passed over the HyperSep solid phase extraction aminopropyl cartridge (200 mg per 3 ml) (60108-425, Thermo Fisher Scientific), which was initially equilibrated with 50 mM acetic acid in 50% methanol three times. Next, the bound compounds were eluted with 600 µl of 500 mM triethylammonium bicarbonate buffer (pH 8.5) in 50% methanol. Then, 600 μl of eluates was dried in the liquid N2 oxidation system with the lids open for 1.5 h. The samples were then solubilized in 1 ml of 10 mM ammonium bicarbonate (pH 8.0), vortexed and passed over the graphite carbon Supelclean Envi-Carb 1 ml column (57109-U, Millipore Sigma) which was pre-equilibrated with 80% acetonitrile + 0.1% formic acid. Next, the bound compounds were eluted with 800 µl of 30% acetonitrile + 10 mM ammonium bicarbonate. Eluate was dried in the liquid N2 oxidation system with the lids open for 2 h. The dried pellets were then frozen in −20 °C.
UHPLC–MS/MS analysis
The concentration of ADP-heptose was determined using an ultra-high-performance liquid chromatography–electrospray ionization MS (UHPLC–ESI-MS/MS) method by modifying our previously described protocol75. A linear calibration curve was generated in the range of 1–1,000 ng ml−1 and 13C6-UDP-glucose (CLM-10513-0.001; Cambridge Isotope Laboratories) was used as an internal standard throughout the assay. The internal standard (10 µl of a 50 ng µl−1 methanol solution) was added to the bacterial lysate samples and to the calibrators and quality-control samples. The 5 µl volume of sample extract was injected on column for analysis by electrospray ionization UHPLC–MS/MS using a Waters TQ-XS triple quadruple mass spectrometer interfaced with an Equity UPLC system. The optimal signals for the ion pair of analyte and internal standard, that is, m/z 617.8 to 270.7 for ADP-heptose and m/z 570.6 to 322.7 for 13C6-UDP-glucose, respectively, were achieved in negative-ion mode with the use of the following instrument settings: capillary voltage, 3.0 kV; source temperature, 120 °C; desolvation temperature, 350 °C; desolvation gas flow, 800 l h−1; and cone gas flow, 150 l h−1. The cone voltage, collision energy and ion dwell time were optimized and were set at 30 V, 30 ev, 0.1 s respectively; helium was used as the collision gas. An ACQUITY UPLC BEH Amide column (2.1 mm × 100 mm, 1.7 µm) was used in separation. A gradient mobile phase was used with a binary solvent system, which started with 25% solvent A and was held for 1 min, changed from 25% solvent A to 100% solvent A over 4 min, held for 2 min, then to 25% solvent A at 7.1 min, and this was held for 3 min. The total run time was 10 min, and the flow rate was 0.2 ml min−1. Solvent A consisted of acetonitrile:water (5:95) with 20 mM ammonium acetate and was adjusted to pH 9.5 with ammonium hydroxide; solvent B consisted of acetonitrile. The injection volume was 5 μl. Data were acquired and processed using Masslynx v.4.1 (Waters).
Faecal microbiota transplantation
To prepare faecal material for transplantation, previously established protocols were followed76,77, and the samples were processed within 30 min of collection. Fresh faecal samples (6 pellets per mouse) were collected from either H2O-treated or DSS-treated mice in 2 ml homogenizer-compatible tubes in 1.5 ml of sterile PBS containing 0.05% l-cysteine HCl reducing agent to preserve anaerobes. After vigorous homogenization at high speed twice to confirm proper mixing, the faecal suspension was filtered through a 40 µM cell strainer to clear away the particulate matter. To further clear out undissolved solids matter and concentrate bacteria, the tubes were centrifuged at 800g for 3 min at 4 °C and the supernatant was collected and diluted with 4.5 ml transfer buffer (1:3). Then, 1 ml aliquots were prepared, stored in 10% glycerol at −80 °C, and used for experiment within 2 weeks of collection. Chimeric mice, as mentioned before, were pretreated with antibiotics cocktail for 4 weeks and then transplanted with faecal material for 4 weeks (twice a week oral gastric gavage 100 µl; every Tuesday and Thursday) after which flow cytometry was performed on BM. To confirm whether faecal microbial transplantation was successful, faecal samples were collected from the donor (H2O- and DSS-treated wild-type) and recipient (Dnmt3a wild-type and knockout) mice at serial timepoints after transplantation (days 3, 7 and 14). DNA was extracted and 16S rRNA-seq was performed to analyse the taxonomic classification of the colonized bacteria.
16S rRNA-seq and analysis
16S rRNA library preparation and metagenomic sequencing was performed using a previously defined protocol. Sequencing was performed using the primer set, 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT), which covers the V4 region to run as PE250 on the MiSeq platform. Read pairs from the raw sequencing data were demultiplexed based on barcodes and downstream data processes were done using USEARCH/UPARSE v.11.0.667_i86linux32 (https://www.drive5.com/usearch)78. In brief, forward and reverse reads were first merged using -fastq_merge pairs, primers were striped (-stripleft 19 -stripright 20) using -fastx_truncate. As low-quality reads often cause spurious OUTs, reads were filtered using -fastq_filter to discard reads with expected error scores below 1. After filtering, the reads were dereplicated with -fastx_uniques. Unique reads were used as input for the uparse step, using -cluster_otus. The -cluster_otus command performs 97% operational taxonomic unit (OTU) clustering, and removes chimeric sequences. The resulting OTU table was normalized to 5,000 reads using -otutab_norm. The OTU tree was also generated using -otutab_norm and -cluster_agg commands. Error-corrected reads (ZOTU, denoised OTU) were identified using the denoising step (-unoise2 command) For taxonomic classification of the bacterial ZOTUs, the -sintax command was used with the reference training set RDP training set v18 (rdp_16s_v18, https://www.drive5.com/usearch/manual/sintax_downloads.html). All non-bacterial ZOTUs were removed on the basis of their taxonomic classification. Microbiome communities in comparison groups were analysed using the R package phyloseq (https://joey711.github.io/phyloseq/). The ZOTU table, the ZOTU taxonomy, the ZOTU tree and the sample table were imported into phyloseq to create the phyloseq object. The possible contaminating DNA features were statistically identified using the decontam package (https://bioconductor.org/packages/release/bioc/html/decontam.html) and the sequenced DNA concentration (efficient ng μl−1) data for each sample. Samples showing signs of substantial contamination were removed at this stage. Statistical analysis of the number of reads, length and mean quality (phred) score were verified using FastQC (v.0.11.8). Example quality scores across the entire read length are presented in Supplementary Table 16. A score of >30 is considered to be very good quality 16S sequence reads. The mean number of reads across all human samples (n = 32) was 131,706 with mean phred Q score of 35.8 and, across mouse samples (n = 78), the mean read depth was 188,177 with mean Q score of 35.7. Any sample with normalized ZOTU < 5,000 reads was not included in the final data analysis. Alpha diversity metrics were computed using the R package vegan (functions diversity, estimate and spec number for Shannon indicator, Chao1 index and observed richness, respectively). Taxonomic classification was then finally investigated. All of the 16S rRNA-seq data generated in this study are provided in Supplementary Tables 17–20 and are available under BioProject ID PRJNA1055136.
Publicly available datasets
RNA-seq data of individuals with AML79 were downloaded from the GDC Data Portal (https://portal.gdc.cancer.gov/) and the BEAT AML (Vizome, http://www.vizome.org/aml/)80. Published microarray data of individuals with MDS81 and respective age-matched controls were downloaded from the GEO (GSE58831). DNA methylation data of wild-type and Dnmt3a−/− HSCs were obtained from the GEO (GSE98191)23.
Statistical analysis
No statistical methods were used to predetermine sample size. The number of animals, cells and experimental/biological replicates can be found in the figure legends. Differences among multiple groups were assessed using one-way and two-way ANOVA followed by multiple-comparison post-testing for all possible combinations. Comparison of two groups was performed using Mann–Whitney U-tests or Student’s t-tests (unpaired, two-tailed) when the sample size allowed. Unless otherwise specified, the results are depicted as the mean ± s.d. or mean ± s.e.m. A normal distribution of data was assessed for datasets > 30. For correlation analysis, the Pearson correlation coefficient (r) was calculated. For Kaplan–Meier analysis, Mante–Cox tests were used. All graphs and analysis were generated using GraphPad Prism 9.0 software or using the package ggplot2 from R. For all analyses, P < 0.05 was considered to be statistically significant. Investigators were not blinded to the different groups. We used the publicly available relative-risk calculator (https://www.gigacalculator.com/calculators/relative-risk-calculator.php) to compute the relative risk (risk ratio), confidence intervals and P values for risk assessment between individuals with CHIP/clonal cytopenias of unknown significance with ADP-heptose present and those with ADP-heptose absent in circulation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.