Mouse strains
Ptf1a-Cre, Ptf1a-Cre;lsl-KrasG12D and Ptf1a-Cre;lsl-KrasG12D;Trp53R172H/+ mice, which were used for the data in Figs. 1a,b,d–h and 2d–f and Extended Data Figs. 1g–k, 2a–1 and 3d,e, and Pdx1-Cre;lsl-KrasG12D;p53R172H/+ mice, which were used for Fig. 4i,j and Extended Data Fig. 12n–s, have been described previously47,48,49. Conditionally floxed Pikfyve (Pikfyvef/f) mice were purchased from Jackson Laboratories. PCR genotyping was done for Ptf1a-Cre mice (for the Ptf1a-Cre, KrasG12D, Trp53R172H/+ and Pikfyvef/f alleles) from DNA isolated from mouse tails using standard methodology. PCR genotyping was done for Pdx1-Cre mice (for the Pdx1-Cre, KrasG12D and Trp53R172H/+ alleles) from DNA isolated from an ear punch using standard methodology. Littermate controls were systematically used in all experiments, and the sex ratios for each cohort were balanced. The Pdx1-Cre animals used for the autochthonous model studies were bred and studied at the CRUK Scotland Institute as previously described50. All experiments involving the Pdx1-Cre animals were approved by the University of Glasgow Animal Welfare and Ethical Review Board and were performed under a UK Home Office licence. All other animals used in this study were housed at the University of Michigan in a pathogen-free environment, and all procedures involving these animals were performed in accordance with requirements of the University of Michigan Institutional Animal Care & Use Committee (IACUC). Mice were housed at a maximum of five mice per cage in a pathogen-free animal facility with a 12 h:12 h light:dark cycle, with 30–70% humidity and a temperature of 20–23 °C.
Cell lines, antibodies and compounds
The cell lines PANC-1, MIA PaCa-2, Panc 04.03, SW1990, Panc 10.05 and HPAF-II were originally obtained from the American Type Culture Collection, and 7940B was provided by Greggory Beatty at Perlman School of Medicine at the University of Pennsylvania. The iKRAS 9805 cell line has previously been described51. The UM PDAC primary cell lines (UM2 and UM19) were obtained from surgically resected samples and established through mouse xenograft52. KPC1344 and KPC1361 were derived from a KPC mouse in-house by dissociating tumours manually with a sterile blade and then treating them with 1 mg ml−1 collagenase II (ThermoFisher Scientific, 17101-015) and 1 mg ml−1 DNase (Sigma-Aldrich, 10104159001) for 30 min with shaking at 37 °C. The cells were then strained using a MACS SmartStrainer (30 μm) (Miltenyi Biotec, 130-110-915) and rinsed with PBS before culturing. The KPC1361-sgNC, KPC1361-sgATG5 and KPC1361-sgATG7 monoclonal lines were generated previously30. All cells were grown and treated in DMEM (Gibco, 12430) + 10% FBS (ThermoFisher) unless otherwise indicated. All cell lines were genotyped to confirm their identity by Eurofins Genomics and tested biweekly for mycoplasma contamination. The sources of all antibodies and compounds are described in Supplementary Table 2. The synthesis and characterization methods used for PIK5-33d are described in Supplementary File 1.
Histopathological analyses
The study pathologists conducted a detailed histopathological evaluation of mouse pancreatic tissues on 4-μm-thick H&E-stained formalin-fixed, paraffin-embedded sections. The examination involved checking all collected pancreas samples for the percentage prevalence of normal pancreas, PanIN, either high or low grade, and lesions with atypia or clear evidence of pancreatic ductal adenocarcinoma. The samples were then classified under these three categories and the results tabulated. Finally, the two pathologists reached a consensus to determine the final percentage prevalence.
PIKFYVE RNAScope
RNA-ISH was performed using an RNAscope 2.5 HD Brown kit (Advanced Cell Diagnostics) and a target probe against PIKFYVE (1326631 Hs-PIKFYVE), according to the manufacturer’s instructions. RNA quality was evaluated in each case using a positive control probe against human housekeeping peptidylprolyl isomerase B (PPIB; 313901). The assay background was monitored using a negative control probe against bacillus bacterial gene DapB (310043). Stained slides were evaluated under a light microscope at low- and high-power magnification for RNA-ISH signals in the cancer cells and normal pancreas by authors R.M. and J.H. The expression level was evaluated according to the RNAscope scoring criteria: a score of 0 means no staining or less than 1 dot per 10 cells; 1 means 1–3 dots per cell; 2 means 4–9 dots per cell, and no or very few dot clusters; 3 means10–15 dots per cell and less than 10% of dots in clusters; 4 means more than 15 dots per cell and more than 10% of dots in clusters. The RNA-ISH score was calculated for each examined tissue section as the sum of the percentage of cells with scores of 0–4 [(A% × 0) + (B% × 1) + (C% × 2) + (D% × 3) + (E% × 4), A + B + C + D + E = 100], using previously published scoring criteria53.
Pikfyve BaseScope
The BaseScope VS reagent kit (Advanced Cell Diagnostics, 323700), which identifies short targets and splice variants, was used to demonstrate Pikfyve on whole-mouse pancreatic tissues. The reagent kit was used with Discovery Ultra automated IHC/ISH slide staining systems by Ventana Medical Systems on a validated protocol utilizing BaseScope VS detection reagents (323710), RNAscope Universal VS sample preparation Reagents v.2 (PN323740) and RNAscope VS accessory kit (320630). BaseScope VS probe BA-Mm-Pikfyve-E6-3zz-st-C1, Mus musculus phosphoinositide kinase FYVE type zinc-finger-containing (PIKfyve) transcript variant 2 mRNA targeting exon 6 complimentary to the target mRNA was used (1300097-C1, accession code NM_011086.2, nucleotides 633–771) for the assay as a test probe. BaseScope VS positive control probe, Mm-PPIB-3ZZ, M. musculus PPIB mRNA (No701079) and BaseScope VS negative control probe DapB-3ZZ (701019) were used as positive and negative controls, respectively.
All slides were examined for positive signals in lesions and background benign pancreas by R.M. and J.H. The RNA in situ hybridization signal was identified as red punctate dots and the expression level was scored as follows: 0 means no staining or less than 1 dot per 10 cells (at ×40 magnification); 1 means 1 dot per cell (visible at ×20 or ×40); 2 means 2–3 dots per cell; 3 means 4–10 dots per cell (less than 10% in dot clusters) visible at ×20; and 4 means more than 10 dots per cell (more than 10% in dot clusters) visible at ×20. A cumulative RNA ISH product score (BaseScope score) was calculated for each available tissue core as the sum of the individual products of the expression level (0–4) and percentage of cells [0–100; that is, (A% × 0) + (B% × 1) + (C% × 2) + (D% × 3) + (E% × 4); total range, 0–400].
Immunohistochemistry
Immunohistochemistry was done on formalin-fixed, paraffin-embedded 4 μm sections of mouse or xenograft tissues. Slides were deparaffinized in xylene, followed by serial hydration steps in ethanol (100%, 95% and 70%) and water for 4 min each. Antigen retrieval was done by boiling slides in citrate buffer (pH 6). Endogenous tissue peroxidase activity was blocked by 3% H2O2 for 1 h. Slides were blocked in 10% goat serum for 1 h. The slides were then incubated in the primary antibodies. The specifics of the antibodies used are listed in Supplementary Table 2. Visualization of staining was done according to the manufacturer’s protocol (Vector Laboratories, SK-4100). Following DAB staining, slides were dehydrated in ethanol (70%, 95% and 100%, 6 min each), xylene (15 min) and mounted using EcoMount (Thermo Fisher, EM897L).
After IHC staining, quantification was done using Fiji (Imagej)54 (Extended Data Fig. 12g). Images were first subjected to colour deconvolution using the H DAB vector. Subsequently, a manual threshold was set on the basis of the uniform signal intensity of the DAB signal, serving as a cut-off for all images. The ratio of brown signal to total signal was calculated as the percentage CK19-positive area displayed on the figure. Regions outside the pancreas, such as the spleen, were excluded from the analysis.
In vivo tumour studies
All animal experiments were done in accordance with the Office of Laboratory Animal Welfare and approved by the University of Michigan IACUC, the CRUK Scotland Institute, or the University of Glasgow. No inclusion or exclusion criteria were used. Both male and female mice were used. Mice were randomized to treatment groups. Sample sizes were determined by preliminary studies and the level of observed effect. Study designs, sample sizes, outcome measures, statistical methods and results are stated in the relevant figure legends. Tumours were measured using digital calipers (two or three times per week) or ultrasound (for the autochthonous model, once a week) in a blinded manner. No experiment exceeded the end points predetermined by IACUC: subcutaneous tumours exceed 2 cm in any direction; tumour ulcerates more than half of its surface area; ulceration that has effusion, appears infected or has haemorrhage; or tumour develops in an area that impairs normal movement or physiological behaviour.
Subcutaneous tumour studies
For xenograft studies, CB17 severe combined immunodeficiency (SCID) mice 6–8 weeks old were obtained from the University of Michigan breeding colony. For syngeneic studies, C57BL/6J mice 6–8 weeks old were obtained from Jackson Laboratories. Subcutaneous tumours were established at both sides of the dorsal flank of the mice by injecting 1 × 106 cells in 100 μl of 50:50 Matrigel and serum-free medium. Tumours were measured 2–3 times per week using digital calipers following the formula (π/6)(L × W2), where L is the length and W is the width of the tumour. At the end of the studies, the mice were killed and tumours were extracted and weighed.
Pancreatic orthotopic tumour studies
The 7940B and UM19 orthotopic models were established according to previously described protocols13. In brief, 50,000 (7940B) or 1,000,000 (UM19) cells were implanted directly into the pancreas of C57BL/6J mice (for 7940B) (Jackson Laboratories) or CB17 SCID (for UM19) mice. Tumours were established for 11 days before treatment with the indicated conditions. Mice were killed after three weeks of treatment, and tumours were weighed and preserved for further analyses.
KPC autochthonous model
The animals used for the autochthonous model studies were bred and were studied at the CRUK Scotland Institute, as previously described50. All experiments were approved by the University of Glasgow Animal Welfare and Ethical Review Board and were performed under a UK Home Office licence.
In vivo apoptosis evaluation using TUNEL staining
TUNEL staining was performed with an in situ cell death detection kit (Roche Applied Science TMR Red, 12156792910) following the manufacturer’s instructions. In brief, fixed sections were deparaffined, rehydrated and subsequently permeabilized using proteinase K. The labelling reaction was done at 37 °C for 1 h by addition of the reaction buffer containing the enzymes. Images were acquired using a Zeiss Axiolmager M1 microscope. Quantification was performed using Fiji (ImageJ)54 (Extended Data Fig. 3r). Signals from TUNEL and DAPI were quantified independently using the same manual threshold for all samples. The TUNEL percentage of positive scores was calculated as a percentage of the TUNEL signal divided by the DAPI signal.
Immunoblots
Cell lysates were prepared in RIPA buffer (ThermoFisher Scientific) supplemented with Halt protease and phosphatase inhibitor cocktail (ThermoFisher Scientific). Total protein was measured by DC Protein Assay Kit II (BIO-RAD). An equal amount of protein was resolved in NuPAGE 3–8%, Tris-acetate protein gel (ThermoFisher Scientific) or NuPAGE 4–12% Bis-Tris protein gel (ThermoFisher Scientific), blocked with 5% non-fat dry milk in TBS-T and blotted with primary antibodies overnight. After incubation with HRP-conjugated secondary antibodies, membranes were imaged on an Odyssey Fc Imager (LiCOR Biosciences). For immunoblot experiments involving multiple targets overlapping in size, sample lysates were prepared in bulk and loaded on multiple gels as needed. Alternatively, membranes were stripped using Restore western blot stripping buffer (ThermoFisher Scientific) according to the manufacturer’s instructions, rinsed, blocked and re-probed. One representative loading control for each experiment was displayed in the figures. For gel source data, see Supplementary Fig. 1.
CETSA
CETSA was done according to previously described protocols55. In brief, 7940B cells were seeded overnight and subsequently treated with DMSO, ESK981 (1,000 nM) or apilimod (1,000 nM) for 2 h. The cells were then collected and used in single-cell suspensions of 1 × 106 cells in 50 μl PBS containing protease inhibitors. The suspensions were then heated and cooled (two cycles of 3-min heating followed by 3-min cooling at room temperature) using a thermal cycler. Cells were then lysed with three cycles of freeze–thawing in liquid nitrogen. Lysates were then centrifuged at 12,000g for 10 min and the soluble fraction was isolated, denatured and resolved on a NuPAGE 4–12%, Bis-Tris protein gel (ThermoFisher Scientific), blocked with 5% non-fat dry milk in TBS-T and blotted with primary antibodies overnight. After incubation with HRP-conjugated secondary antibodies, membranes were imaged on an Odyssey Fc Imager (LiCOR Biosciences).
Radioactive inositol labelling and measurement of phosphorylated phosphoinositide lipids
Radioactive phosphoinositide lipid labelling was done as previously described56. In brief, PANC-1 cells were grown in 100-mm dishes to 70–80% confluence. Cells were rinsed twice with 1 × PBS, pH 7.4, and were incubated in inositol-free medium with 10 µCi ml−1 [3H]inositol for 24 h. The cells were then treated as indicated in Fig. 2b,c, Extended Data Fig. 3a–c or Extended Data Fig. 4i–m at a final concentration of 1 µM and incubated for the indicated time. Note that the small molecules were added into the labelling medium at the beginning of the labelling procedure for the 24-h samples. Following the treatments, myo-[2-3H]inositol-labelled lipids were extracted as described56 and the resultant glycerophosphoinositides were separated by ion exchange chromatography on a Partisphere 5 μm SAX cartridge column, 250 × 4.6 mm (WVS Hardware, 4621-1505, MAC-MOD Analytical). The raw counts for each peak are presented as a percentage of the total phosphatidylinositol, which is derived from the summation of counts across the six detectable glycero-inositol peaks (PtdIns, PtdIns3P, PtdIns4P, PtdIns5P, PtdIns(3,5)P2 and PtdIns(4,5)P2). Background scintillation counts, determined from adjacent regions, were subtracted from all peaks.
Cell viability assays and synergy assays
Cells were plated into 96-well plates and incubated overnight at 37 °C in 5% CO2. The next day, a serial dilution of the indicated compounds was prepared in culture medium and added to the plate. The cells were then further incubated for five days for experiments involving MRTX1133 or trametinib, or seven days for all other experiments. Subsequently, a CellTiter-Glo assay (Promega) was then done according to the manufacturer’s instructions. The luminescence signal was acquired using an Infinite M1000 Pro plate reader (Tecan) and the data were analysed using GraphPad Prism 10 (GraphPad Software).
To determine the synergism of two different compounds using viability assays, cells were treated with the indicated combinations of the drugs for five days before we did the CellTiter-Glo assay as described above. These experiments were done with five biological replicates, each with ten wells of untreated internal controls for each plate used in each experiment, which were used for normalization between plates. The data were then expressed as percentage inhibition relative to baseline, and the presence of synergy was determined by the Bliss method using the SynergyFinder+ web application57.
Autophagic flux probe generation and assay
Generation of the autophagic flux probe in 7940B, Panc 04.03 and iKRAS 8905 cells was done according to the original author’s instructions35. In brief, cells were infected with pMRX-IP-GFP-LC3-RFP-LC3ΔG, which was a gift from Noboru Mizushima (Addgene, 84572). After puromycin selection, single-cell clones were expanded and genotyped to ensure the absence of homologous recombination between two LC3 fragments during retrovirus integration.
We seeded 15,000 cells in 96-well plates. After overnight incubation, cells were treated with the indicated compounds for 24 h. For assays assessing the co-treatment of autophagy inhibitors (apilimod, ESK981 or chloroquine) with autophagy inducers (torin-1, trametinib or MRTX1133), the autophagy inhibitor was added 4 h before the inducer. For assays using iKRAS 8905, cells were seeded with or without doxycycline, as indicated, and then treated with compounds in a similar fashion. Fluorescent signals were detected using the Infinite M1000 Pro plate reader (Tecan). Autophagy index was calculated by dividing the RFP signal by the GFP signal from each well, followed by normalization of all RFP/GFP ratios by the average RFP/GFP ratio of the DMSO condition.
Confluence-based proliferation assays
Cells were seeded in a clear 96-well plate overnight before treatment. For treatment with the indicated compounds, plates were incubated in an Incucyte S3 2022 Rev1 (Sartorious), with ×10 images taken every 4 h, and confluence was analysed to assess for proliferation.
Oxygen-consumption assays
The OCRs were determined using a Seahorse XF glycolytic rate assay (Agilent), according to the manufacturer’s protocol. In brief, 15,000 (7940B) or 25,000 (Panc 04.03) cells were seeded in an Agilent XF96 cell culture microplate 16 h before treatment. Cells were treated with apilimod, ESK981, chloroquine or bafilomycin as indicated for 8 h. Immediately before the assay, cells were washed and then incubated in XF DMEM medium (pH 7.4, Agilent) with 1 mM pyruvate, 2 mM glutamine and 10 mM glucose. The assay was done on an XF96 extracellular flux analyser (Agilent), and the OCR was calculated using Seahorse Wave Controller software (v.2.6.3.5, Agilent). The OCR was normalized to cell number using the CyQUANT NF cell proliferation assay (Invitrogen) according to the manufacturer’s instructions.
Real-time monitoring of basal OCR was done using a Resipher (Lucid Scientific). Next, 15,000 7940B cells were seeded in 50 μl of medium in a clear 96-well plate 16 h before treatment. Immediately after treatment with an additional 50 μl of medium (for a total of 100 μl), OCR monitoring was started by placing the Resipher device on the cells, which were incubated at 37 °C and 5% CO2 for 24 h.
Metabolic CRISPR screen
The human CRISPR metabolic gene knockout library was a gift from David Sabatini (Addgene, 110066)58. To achieve at least 1,000-fold coverage of the library for culturing, 75 × 106 MIA PaCa-2 cells were seeded at a density of 5 × 105 cells per ml in six-well plates containing 2 ml DMEM, 8 mg ml−1 polybrene and the CRISPR screen library virus. Spin infection was done by centrifugation at 1,200g for 45 min at 37 °C. After incubation for 24 h, the medium was replaced with fresh DMEM. After a subsequent 24-h incubation, cells were transferred to T-150 flasks (at a density of three wells into one T150 flask) containing 20 ml DMEM with puromycin at 2 mg ml−1. After three days of selection, cells were seeded into 16 T-150 flasks at a density of 5 x 106 cells per flask in 20 ml DMEM containing either DMSO or 100 nM apilimod. Cells were passaged every 3–4 days and reseeded back to the original cell density before collection on day 14. For the high-dose CRISPR screen, 2 μM apilimod was used and was refreshed every 3 days until cells became confluent at day 17 and were collected. We collected 15 million cells from each condition for isolation of genomic DNA using the DNeasy blood and tissue kit (Qiagen), according to the manufacturer’s protocol.
For each condition, sgRNA was amplified from 50 mg genomic DNA using Herculase II fusion DNA polymerase (Agilent Technologies), column purified using Select-a-Size DNA Clean & Concentrator kit (Zymo Research) and then gel-purified using 6% Novex TBE gel (Thermo Fisher), followed by isolation from the gels with gel breaker tubes and gel filters (BioChain). The resulting PCR products then underwent end-repair and A-tail addition followed by New England Biolabs (NEB) adapter ligation. The final library was prepared by enriching adapter-ligated DNA fragments using 2 × KAPA HiFi HotStart mix and NEB dual-code barcode following the manufacturer’s protocol. The libraries were then sequenced on an Illumina NovaSeq 6000 (paired-end 2 × 151 nucleotide read length).
Reads were trimmed to the bare sgRNA sequence using cutadapt 4.1 (ref. 59). Paired-end mates were trimmed separately using a sequence 5′-adjacent to the sgRNA position within the vector (TATATCTTGTGGAAAGGACGAAACACCG), requiring a minimum match of 18 bases to the sequence and followed by truncation to 20 bases (relevant cutadapt command parameters: -m 18 -O 18 -l 20 –discard-untrimmed). Trimmed reads were then combined and aligned using bowtie2 2.4.5 (ref. 60) to a reference built from each sgRNA in the library flanked by vector sequences (5′-GTTATCAACTTGAAAAAGTGGCACCG and 3′-CTAGATCTTGAGACAAATGGC). The bowtie2 parameter –norc was used to prevent reverse compliment alignment. Counting was then done using MAGeCK 0.5.9.5 (ref. 61). Supplementary Table 1 contains a summary of read counts. sgRNAs with fewer than 100 counts in the initial dataset were removed from downstream analysis. Genes targeted by fewer than six distinct sgRNAs following this filtering were likewise removed. Downstream analyses, including calculation of sgRNA depletion/enrichment scores, gene depletion/enrichment scores and selective dependency, were done according to previously described methods62. In brief, normalized sgRNA abundances were calculated by adding a pseudocount of one and then normalized to the total counts of each sample. The sgRNA enrichment/depletion scores were calculated as log2 fold change between the final and initial populations, and the gene scores were calculated as the average log2 fold change of the sgRNAs targeting that gene. To calculate selective essentiality scores, we first scaled gene scores using the medians of non-targeting sgRNAs and sgRNAs targeting core essential genes as references (0 and −1, respectively). Selective essential genes were then identified by taking the z-scored difference between the scaled apilimod and DMSO gene scores. Plots were generated using ggplot2 (v.3.4.4).
RNA isolation and qPCR
Total RNA was isolated from cells using the miRNeasy kit (Qiagen) or DirectZol RNA Miniprep kits (Zymo), and cDNA was synthesized from 1,000 ng of total RNA using a Maxima First Strand cDNA synthesis kit for reverse transcription (RT)-qPCR (Thermo Fisher Scientific). RT-qPCR was done in triplicates using standard SYBR green reagents and protocols on a QuantStudio 5, 6 pro or 7 Real-Time PCR system (Applied Biosystems). The target mRNA expression was quantified using the ΔΔCt method and normalized to ACTB (human) or Actb (mouse) expression. Data presented represent technical triplicates. All primers were synthesized by Integrated DNA Technologies. Primer sequences are listed in Supplementary Table 2.
RNA-seq and analysis
RNA-seq libraries were prepared using 800 ng total RNA. Ribosomal RNA was removed by enzymatic digestion of the specific probe-bound duplex rRNA and then fragmented to around 200–300 base pairs with heat in fragmentation buffer (KAPA RNA Hyper+RiboErase HMR, Roche). Double-stranded cDNA was then synthesized by reverse transcription and underwent end-repair and ligation using New England Biolabs adapters. We did final library preparation by amplification with 2 × KAPA HiFi HotStart mix and NEB dual barcode. Library quality was measured on an Agilent 2100 Bioanalyzer (DNA 1000 chip) for concentration and product size. Paired-end libraries were sequenced using an Illumina NovaSeq 6000, (paired-end 2 × 151-nucleotide read length) with sequence coverage to 30 million to 40 million paired reads. Reads were demultiplexed using Illumina bcl2fastq conversion software (v.2.20). Transcripts were quantified by the alignment-free approach kallisto63 using index generated from mouse reference genome (mm10) and then summed to obtain gene level counts. Raw transcripts per million values for each gene are shown in Supplementary Table 3. Differential analysis was done using limma-voom64,65 after TMM normalization66 of gene level counts with calcNormFactors of edgeR67. Genes with mean transcripts per million of less than 1 in both control and treatment groups were considered as low-expressed genes and excluded from differential analysis. Enrichment of the Hallmark and Reactome gene sets downloaded from MSigDB68 were examined using fgsea69 with genes ranked by logFC estimated from limma as input.
Immunofluorescence
Cells were seeded overnight on an eight-chamber glass slide (CELLTREAT) in 500 µl culture medium. The next day, cells were treated as indicated for each experiment and then fixed using 3.2% paraformaldehyde for 15 min, quenched with 125 mM l-glycine for 10 min and then rinsed twice with PBS. For LAMP1 immunofluorescence, samples were then permeabilized with 0.1% Triton-X 100 for 5 min, rinsed three times with PBS, blocked in 5% BSA for 1 h at room temperature and then incubated in LAMP1 primary antibody (Supplementary Table 2), 1:100 overnight at 4 °C. Samples were rinsed three times with PBS and then incubated in goat anti-rabbit secondary antibody (Alexa Fluor 594), 1:1,000, at room temperature for 1 h. For filipin immunofluorescence, cells were rinsed three times with PBS and incubated in 0.1 mg ml−1 filipin complex for 2 h at room temperature. Samples were then rinsed three times in PBS and mounted with PBS and imaged on a Zeiss LSM900 confocal microscope (filipin, 405 nM channel; LAMP1, 568 nM channel; Airyscan mode) at ×63 magnification.
Chromatin immunoprecipitation followed by sequencing
All the steps of the chromatin immunoprecipitation followed by sequencing (ChIP-seq) experiments were done as previously described70 using the ideal ChIP-seq kit (Diagenode) with the following specifications. First, 5 million MIA PaCa-2 cells were treated as indicated for 8 h. The sonication cycle used was 30 s on followed by 30 s off, easy mode, for four cycles (Biorupter, Diagenode) to achieve an average fragment size of 200 base pairs. We used 4 μg of c-MYC antibody for immunoprecipitation of fragmented chromatin. ChIP DNA was then de-crosslinked, purified and prepared for sequencing, as previously described70.
Libraries were sequenced on a NovaSeq 6000, producing 150-base pair end reads. Reads were trimmed using Trimmomatic 0.39 (ref. 71) with options PE ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10. Trimmed paired reads were aligned to the human reference genome using bwa72 with options -5 -S -P -T 0. The GRCh38/hg38 reference sequence was obtained from UCSC. Alignments were sorted and filtered for mapping quality >= 20 using samtools 1.9 (ref. 73). Read duplicates were removed using Picard MarkDuplicates 2.26.0 (http://broadinstitute.github.io/picard/). Non-primary alignments were removed using samtools view (option -F 0×900) and converted to BED format using bedtools bamtobed 2.27 (ref. 74). Peaks were called from these alignments using MACS2 2.2.7 (ref. 75) with default settings and the -B option to generate bedGraph coverage files. Peaks were then filtered using the ENCODE Unified GRCh38 exclusion list (https://www.encodeproject.org/files/ENCFF356LFX/). Coverages captured by MACS2 were converted to bigWig using wigToBigWig 2.4 (ref. 76). FIMO 5.5.6 from the MEME77 was used to find the JASPAR Myc motif MA0147.3 in called peaks78. Motif enrichment was done using findMotifsGenome.pl (HOMER 5.1 (ref. 79)) with options -size 200 -len 8 on filtered peaks (score >60).
Generation of CRISPRi-mediated knockdown cell lines
The sgRNA sequences used were taken from a previously validated Perturb-seq library80. The sgRNAs were cloned into the backbone pLV hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro (Addgene: plasmid 71236; http://n2t.net/addgene:71236; RRID: Addgene_71236)81 using the Golden Gate reaction. The generated plasmids were then expanded, verified by Sanger sequencing and packaged into lentiviruses by the University of Michigan Vector Core. Cells were seeded, infected with viruses along with polybrene (10 mg ml−1) and then selected with puromycin (2 μg ml−1 for MIA PaCa-2 and 5 μg ml−1 for PANC-1) before further analysis. Given the notable impact of PIKFYVE and FASN knockdown on PDAC cells, new CRISPRi knockdown cell lines were generated before each experiment. The sgRNA sequences are listed in Supplementary Table 2.
Generation of CRISPR-mediated gene knockout cell lines
The sgRNA sequences used are provided in Supplementary Table 2 and were inserted into the lentiCRIPSRv2 backbone82 using the Golden Gate reaction, amplified and verified by Sanger sequencing. Plasmids were transfected into cells using Lipofectamine 3000 (ThermoFisher Scientific) according to manufacturer’s protocol. Cells were then selected using puromycin as described above before further analysis. Of note, to knock out AMPKα, two sgRNAs targeting AMPKα1 (PRKAA1) and 2 sgRNAs targeting AMPKα2 (PRKAA2) were transfected together (625 ng each, totalling 2.5 μg of plasmid).
Knockdown of PRKAA1/PRKAA2 using siRNA
ON-TARGETplus Human SMARTpool siRNA (Horizon) targeting PRKAA1 and PRKAA2 or a non-targeting control were transfected into PANC1 cells using Lipofectamine RNAiMAX (ThermoFisher Scientific) at a concentration of 25 nM according to the manufacturer’s protocol. One day after transfection, cells were trypsinized and re-seeded. After a subsequent overnight incubation, cells underwent a second round of transfection before further analysis.
ESK981, trametinib, selumetinib and MRTX1133 formulation
ESK981 was added to ORA-PLUS and sonicated until completely dissolved. Trametinib was added to corn oil and sonicated until completely dissolved. Aliquots were frozen at −20 °C to prevent freeze–thaw cycles. Selumetinib was suspended in 0.5% hydroxypropyl methylcellulose (HPMC) + 0.1% Tween-80 in water and kept suspended by continuous stirring at 4 °C for up to one week. The MRTX1133 used for the subcutaneous study was added to 10% Captisol in 50 mM citrate (pH 5.0), sonicated until completely dissolved as previously described43, and kept at 4 °C in the dark for a maximum of five days. The MRTX1133 used for the KPC autochthonous model was dissolved in hydroxypropyl-B-cyclodextrin (10% w/v in 50 mM citrate, pH 5.0) and kept with continuous stirring at 4 °C for up to one week. The ESK981, trametinib and selumetinib were delivered by oral gavage. The MRTX1133 was delivered by intraperitoneal injection.
Targeted metabolomics
Polar metabolites from samples treated in biological triplicates were extracted using 80% (v/v) methanol/water and normalized using protein quantification from an additional sample from each condition. Equal estimated amounts of metabolites were dried using a SpeedVac vacuum concentrator, reconstituted in 50% (v/v) methanol in water, and analysed by liquid chromatography–tandem mass spectrometry (LC–MS/MS), as previously described83. Data were analysed as previously described83 using Agilent MassHunter Workstation Quantitative Analysis for QQQ v.10.1, build 10.1.733.0. However, metabolite abundance levels were not divided by the median levels across the samples. No post-detection normalization was done to avoid assuming linearity of signal. Raw values of each metabolite measured are provided in Supplementary Table 4. Heatmaps were generated using the Morpheus Matrix Visualization and analysis tool (https://software.broadinstitute.org/morpheus).
Targeted lipidomics
Experiments with results presented in Extended Data Fig. 7e–h used the following methods.
Sample preparation
Samples for lipidomics analyses were prepared according to the automatic dual-metabolite/lipid sample-preparation workflow described in the Agilent application note 5994-5065EN. In brief, 1 million cells were washed in PBS and lysed with 1:1 trifluoroethanol:water at room temperature. Lysates were transferred to microcentrifuge tubes, incubated for 10 min and centrifuged at 250g for 30 s. Samples were dried with a vacuum concentrator and resuspended in 1:1 trifluoroethanol:water. After transferring the samples to a 96-well plate, lipids were selectively isolated on a Bravo automated liquid handler platform (Agilent) operated by a VWorks automation protocol as described (5994-5065EN).
LC–MS/MS analysis
Samples were analysed on an Agilent 1290 Infinity II Bio LC ultra-high-performance liquid chromatography system with the Agilent Standardized Omics LC configuration, consisting of a high-pressure binary pump, multicolumn thermostat and a temperature-controlled multi-sampler. Samples were injected in randomized order on an Agilent 6495 C triple quadrupole mass spectrometer equipped with an Agilent Jet Stream Dual ESI ion source. Samples were analysed using the reverse-phase LC–MS/MS method reported in the Agilent application note 5994-3747EN. After acquisition, datasets were processed using MassHunter Quantitative Analysis 12.0 software and subsequently imported into Mass Profiler Professional (MPP) for chemometric analysis. No post-detection normalization was done to avoid assuming linearity of signal. Raw values of each lipid measured are provided in Supplementary Table 5.
Experiments with results presented in Extended Data Figs. 7i,j and 11k,l used the following methods.
After cell treatment as indicated, cells were pelleted by centrifugation at 1,000g for 4 min at 4 °C. Next, 1 ml of chilled 10:3 methyl tert-butyl ether:methanol was added to the cell pellets, vortexed for 20 s and incubated at 4 °C for 5 min. Then 188 μl of MS-grade water was added and the sample was vortexed for 20 s. The samples were then centrifuged at 14,000g at room temperature for 2 min, then 700 μl of the lipid fraction (top) was then isolated and moved to a separate tube. Using an independent biological replicate of all conditions, total protein was measured using the DC Protein Assay Kit II (BIO-RAD) as previously described. Using the protein quantity, the volumes of lipid extracts were normalized and then dried.
Dry lipid extracts were reconstructed with 20 μl of 9:1 methanol:chloroform, vortexed for 2 min and centrifuged for 10 min at 13,000 rpm at 20 °C to pellet insoluble material. Supernatants were transferred to analytical vials containing glass inserts and analysed by LC–MS/MS. Lipids were separated by reverse-phase C18 chromatography on an Agilent 1290 Infinity II BioLC with Agilent standard omics configuration coupled with an Agilent 6495D iFunnel triple quadrupole mass spectrometer. Details of the LC–MS/MS acquisition method are reported in the Agilent Application note 5994-3747EN. Samples were injected in randomized order and the raw data were processed using Agilent Mass Hunter Quantitative Analysis 12.0. Lipid signals were exported as CSV files for chemometric analysis. Raw values of each lipid measured are provided in Supplementary Table 5.
Changes in lipid class abundance in 7940B cells following treatment with apilimod (100 nM) or ESK981 (1,000 nM) relative to treatment with DMSO were estimated from linear mixed models with random intercepts to adjust for the baseline differences across the lipid classes. A separate model for each treatment (apilimod or ESK981) comparison against DMSO was built using the R package lme4 (v.1.1-35.1)84.
Stable isotope tracing
Target compound confirmation
C20 ceramide (d18:1/20:0) C38H57NO3 (Cayman Chemicals) and C22 ceramide (d18:1/22:0) C40H79NO3 (Cayman Chemicals) standards were dissolved in methanol at a concentration of 0.1 μg μl−1 and were used to confirm the identity of each species. (M + H)+, (M-H2O + H)+, (M + Na)+, (M + NH4)+ and (M-H)− were used to confirm the identity of each lipid species with mass accuracies of 5 ppm tolerance.
Samples preparation and data analysis
After allowing cells to attach overnight, the culture medium was changed to DMEM − glucose with either 4.5 g l−1 of U-13C6 isotopically labelled glucose or 12C6 glucose, as indicated. Cells were also treated with PIKfyve inhibitors or DMSO as indicated. After 24 h, lipid extracts were isolated using methyl tert-butyl ether following the procedure described above. Protein lysate was extracted and quantified from a separate biological replicate of each condition for normalization. Samples were then normalized, dried and dissolved in 50 μl methanol. A 2 μl sample was then run on a Thermo Scientific IQX Orbitrap LC–MS system with an A YMC Accura Triart C8 (12 nm, 1.9 µm) 150 × 2.1 mm ID Column and a Phenomenex High Pressure column protection filter for separation. Skyline (v.24.1) was used to analyse the ion counts for the mass isotopologues for each species. Raw values of each lipid measured are provided in Supplementary Table 5.
Statement on the use of human samples
Patient tissues from biopsies of pancreatic tumours were acquired from the University of Michigan pathology archives. These tissues were used for RNA Scope (RNA-ISH) experiments to assess for PIKFYVE expression in tumour or adjacent healthy pancreatic cells. The use of clinical formalin-fixed paraffin-embedded specimens from the archives was approved by the University of Michigan Institutional Review Board and did not require patient consent.
Statistical analyses
No statistical methods were used to predetermine sample sizes. For all in vivo experiments, animals were randomly assigned into treatment cohorts. Tumour measurements were done by digital calipers or ultrasound (for the autochthonous model) in a blinded manner. For all in vitro experiments, cells were seeded from the same pool, so there was no requirement for randomization. All samples were analysed equally and simultaneously to eliminate bias. GraphPad Prism software (v.10) and R (v.4.3.2) were used for statistical calculations. Specific R packages used for individual analyses were included in their specific section in the Methods.
Statistics and reproducibility
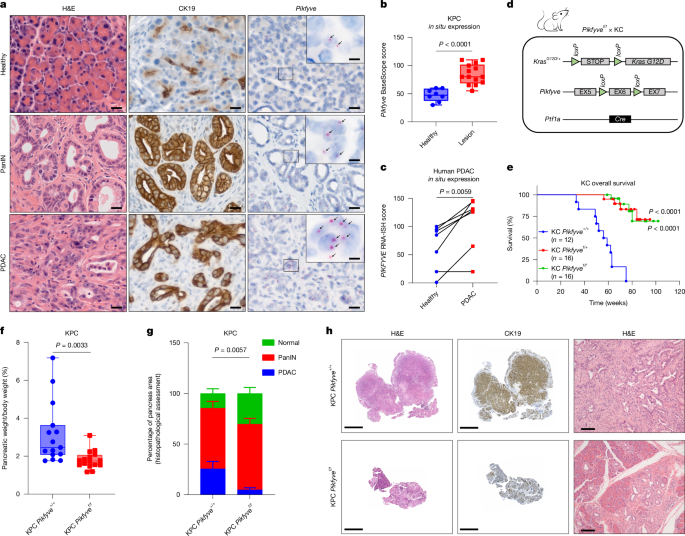
Figure 1. In b, the normal group had n = 8 samples from individual animals and the lesion group had n = 14 samples from individual animals; 8 animals were shared between the two groups. P-value, 3.6 × 10−5. c, Biopsy samples were taken from five independent PDAC patients: two patients donated two samples each from distinct biopsies. Scores were determined as described above. e, P-values: KC Pikfyve+/+ versus KC Pikfyvef/+, P = 7.4 × 10−8; KC Pikfyve+/+ versus KC Pikfyvef/f, P = 1.4 × 10−9. f, n = 15 individual KPC Pikfyve+/+ animals; n = 16 individual KPC Pikfyvef/f animals. g, n = 15 individual KPC Pikfyve+/+ animals; n = 16 individual KPC Pikfyvef/f animals. h, These images are representative of n = 15 KPC Pikfyve+/+ and n = 16 KPC Pikfyvef/f.
Figure 2. a, This experiment was performed once. b,c, One biological replicate of each condition was analysed in two independent experiments for a total of n = 2 for each group. e, Vehicle, n = 11; ESK981, n = 11; WT, n = 8 animals. f, n = 11 individual animals for both groups. g, The data for the ‘no tumour’ group was also used as a reference in Fig. 4d (n = 8 tumours for the vehicle and ESK981 groups, and n = 6 for the ‘no tumour’ group, all from individual animals). P-values: vehicle versus ESK981, 7.4 × 10−5; vehicle versus no tumour, 3.9 × 10—6; ESK981 versus no tumour, 0.19. i, n = 8 animals for each group. j, Vehicle, n = 9; ESK981, n = 9; no tumour, n = 5 animals. k, These images are representative of n = 9 for each group.
Figure 3. f, These images are representative of n = 2 images.
Figure 4. a, These experiments were each performed independently twice with similar results. b, n = 3 technical replicates for each group. These experiments were performed independently three times, each with similar results. P-values: Fasn: ESK981, +Dox versus −Dox, 1.8 × 10−5; Acaca: DMSO, +Dox versus –Dox: 8.8 × 10−5. c, This experiment was performed twice with similar results. d, n = 9 individual animals for vehicle, trametinib and ESK981 groups; n = 8 animals for the ESK981 + trametinib group; n = 6 for the no tumour group. P-values for vehicle versus ESK981 + trametinib, P = 8.4 × 10−6. e, n = 9 animals for vehicle, trametinib and ESK981 groups; n = 8 individual animals for the ESK981 + trametinib group. g, n = 16 tumours from 8 animals for the vehicle group; n = 14 tumours from 7 animals for the trametinib, ESK981 and ESK981 + trametinib groups. P-value: less than 1 × 10−15. h, P-values: ESK981 + trametinib versus vehicle, 7.15 × 10−175; ESK981 + trametinib versus trametinib, 6.7 × 10−137; ESK981 + trametinib versus ESK981, 2.8 × 10−54. i, n = 5 animals for the vehicle group; n = 8 animals for the selumetinib group; n = 6 for ESK981 and ESK981 + selumetinib groups. j, n = 5 individual animals for the vehicle group; n = 8 animals for the selumetinib group; n = 6 for the ESK981 and ESK981 + selumetinib groups.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.