Mice
All experiments with wild-type mice were performed with C57BL/6J mice. All engram-labelling experiments were performed with Fos-tTa17,20, Trap2;Ai32 (refs. 33,48) and Trap2;R26R (refs. 19,34) transgenic mice. Both male and female mice were used. All mice were group housed in a 12:12-h (07:00–19:00) light–dark colony room at 22 °C. Food and water were provided ad libitum and all testing was performed during the light phase. Mice used for experiments were approximately 8–12 weeks of age. All animal work was performed in compliance with the Ryan and Lynch laboratory project licences, with ethical approval of the Trinity College Dublin ethics committee, the Animal Research Ethics Committee from the Health Products Regulatory Authority, and according to the Brigham and Women’s Hospital Institutional Animal Care and Use Committee guidelines.
Metabolic cages
To capture metabolic and behavioural information, mice were singly housed in metabolic cages (Promethion, Sable Systems) for a duration of 8 h per day. Each cage included a ceiling-mounted food hopper, water bottle and body-mass monitor. Mice had ad libitum access to food and water throughout the entire 8 h. Oxygen consumption, carbon dioxide emission, energy expenditure, body weight, food and water intake and locomotor activity were monitored throughout the session. In brief, respiratory gasses were measured with an integrated fuel cell oxygen analyser, a spectrophotometric carbon dioxide analyser and a water vapour pressure analyser (GA3m1, Sable Systems)49. Before each run, gas sensors were calibrated with 100% N2 as zero reference and with a span gas containing a known concentration of 0.933% CO2. Air flow was measured and controlled with the multichannel mass flow generator (FR8-1, Sable Systems). Flow rate was set at 2,000 ml min–1. Oxygen consumption and carbon dioxide production were measured for each mouse at 3–4-min intervals. Energy expenditure was calculated using the equation50: \({K}_{{\rm{cal}}/{\rm{h}}}=60\times (0.0003941\times {V}_{{{\rm{O}}}_{2}}+0.001106\times {V}_{{{\rm{CO}}}_{2}}\).
Cold training schedule
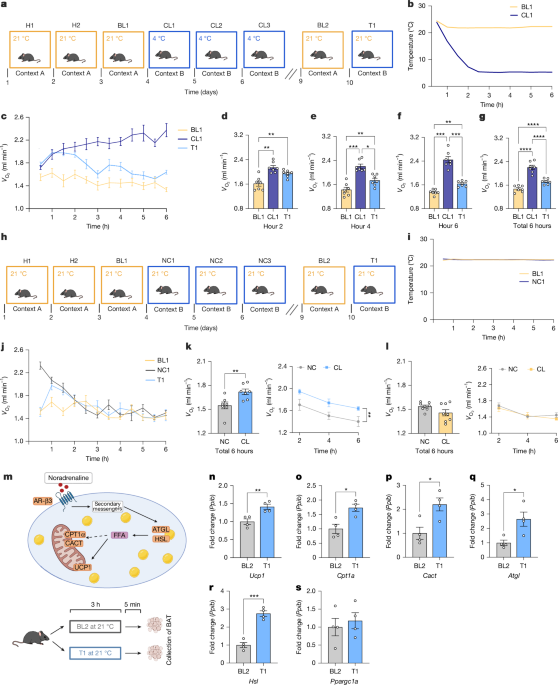
Male mice were taken from their home cage each morning and individually housed in the Promethion cages for a maximum of 8 h. On days 1 and 2, mice were placed in the Promethion cages at 21 °C to allow them to habituate to the machine. On day 3, mice were put in the Promethion cages at 21 °C while metabolic and behavioural data were simultaneously recorded to establish baseline measurements (context A). On day 4, the cages were changed before the mice were placed inside. Contextual cues were added to the chambers, including wall and floor patterns, brighter lights, different bedding and three drops of acetic acid odour (context B). Once the mice were placed in the cages, the temperature decreased in increments of approximately 10 °C per hour. As such, the mice spent a total of 6 h in the altered context at a temperature of 4 °C. This was repeated for a total of three days, with metabolic and behavioural data recorded throughout. Mice were then returned and kept in their home cages for a two-day break period. Next, on day 9, mice were put back in the Promethion cages without any of the additional cues at 21 °C to allow for a second baseline measurement (context A). Finally, on day 10 all the additional context cues were added back in before the mice were put into the cages (context B). Temperature was kept at 21 °C and metabolic and behavioural data were simultaneously recorded to establish the test-day measurements.
Predator odour exposure in metabolic cages
Mice were placed in the metabolic cages 24 h before odour application. In brief, 20 µl of either H2O or 10% w/v trimethylthiazoline were dropped onto a small 1-cm square piece of filter paper in the cage. Mice were exposed to the odorant for 1 h before being put back into their home cage. Metabolic data were recorded at all times.
Conditioned cold aversion
The conditioned place preference apparatus consisted of two separate chambers. Each chamber had different wall and floor cues (black and white pattern). Mice could freely enter both chambers through an opening in the wall in the middle of the apparatus. On day 1, mice were taken from their home cage and placed inside the apparatus, where they could freely explore both sides of the apparatus for a total of 20 min. The entire apparatus was kept at 21 °C. On day 2, the opening in the middle wall of the apparatus was closed. During the morning sessions, mice were exposed to one side of the apparatus at 21 °C for a duration of 2 h. Mice were then returned to their home cage for a 2-h break. In the afternoon sessions, mice were then exposed to the other side of the apparatus at 4 °C for a duration of 2 h. This was subsequently repeated for a total of three days. On day 5, the opening in the middle wall of the apparatus was re-opened before mice were placed back in the apparatus. Mice could freely explore both chambers again for a total of 20 min while the entire apparatus was kept at 21 °C. All behaviours were videotaped and subsequently hand scored. Time spent in each chamber in seconds was measured on day 1 (pre-test) and day 5 (test). The time spent in each chamber was normalized using the formula (test phase duration spent in the cold-paired chamber/time spent there in the pre-test phase), to account for any pre-existing preferences51. Both time of day and side of apparatus were counterbalanced across all mice.
Contextual fear conditioning
Before training, mice were handled for three consecutive days to habituate them to the experimenter. One day before the training day, mice were habituated to the arena and allowed to freely explore the context for 10 min. The next day, mice underwent a contextual fear conditioning protocol. The mice were allowed to explore the arena for the first 3 min. After these 3 min, half of the mice received three foot shocks (0.2 s, 0.75 mA) that were spaced by intervals of 1 min, whereas the other half of the mice received no foot shocks. Mice were subsequently returned to their home cage for 45 min.
Predator odour exposure for BAT collection
Mice were placed into a vacuum-powered four-chamber olfactory arena, which was replicated from previously published designs52. Air entered each corner of the arena at a flow rate of 150 ml min–1 and was odorized by passing through a bottle containing 20 ml of H2O or 350 mM trimethylthiazoline. Mice were exposed to the odour for 5 min and were then euthanized for tissue collection.
Tissue collection
Cold memory
Brains and BAT were collected from mice that went through the above-described cold memory timeline. Tissues were collected on BL1, CL1, BL2 and T1. To ensure that the differences seen were due to the differences in conditions and not time of day, all tissues were collected exactly 5 h after the mice were put inside the Promethion cages.
Contextual fear conditioning
For the contextual fear conditioning experiments, brains and BAT were collected 45 min after the training session ended.
Predator odour exposure
For the predator odour exposure experiments, BAT was collected exactly 1 h after the mice were exposed to the odours.
Tissue preparation
Brains
Mice were transcardially perfused with 4% paraformaldehyde and brains were fixed in 4% paraformaldehyde overnight. The next day, brains were transferred into phosphate-buffered saline (PBS) and kept at 4 °C. They were subsequently sliced in 100-μm coronal sections with a vibratome (Leica VT1200 S) and collected in PBS.
BAT
Mice were euthanized by carbon dioxide inhalation and adipose tissue was removed by making a careful incision between the shoulder blades. Interscapular BAT was subsequently collected, snap frozen in liquid nitrogen and stored at −80 °C.
Immunohistochemistry
To get clear immunolabelling, we used the previously described iDISCO-based immunohistochemistry protocol53,54. Sections were rinsed three times in 1× PBS before being dehydrated in 50% MeOH/PBS at room temperature for 2.5 h. Sections were then rinsed three times in 0.2% PBS with Triton X-100 and blocked in 0.2% PBS with Triton X-100 containing 10% dimethyl sulfoxide and 6% normal goat serum for 2 h at room temperature. After blocking, slides were washed three times in PBS with 0.2% Tween-20 and 10 μg ml–1 heparin (PTwH). Sections were then incubated with primary antibodies (chicken polyclonal anti-GFP (1:1,000, Invitrogen) and rabbit polyclonal IgG anti-FOS (1:1,000, Synaptic Systems)) at 4 °C on a shaker for three days in PTwH with 5% dimethyl sulfoxide and 3% normal goat serum. On day 4, sections were washed three times in PTwH and incubated with secondary antibodies (Alexa Fluor 568 anti-rabbit IgG (1:500, Invitrogen) and Alexa Fluor 488 anti-chicken IgG (1:500, Invitrogen)) overnight at 4 °C. The next day, sections were washed three times in PTwH, followed by three washes in PBS at room temperature. Finally, sections were stained with DAPI in PBS for 10 min followed by a final wash in PBS. Sections were then mounted on superfrost slides using Vectashield DAPI, cover-slipped and sealed.
Confocal imaging
All images were taken on a confocal scanning microscope (Leica TCS SP8, Leica Microsystems). Fluorescence from DAPI was detected at 417–488 nm, Alexa Fluor 568 was detected at 595–650 nm and Alexa Fluor 488 was detected at 500–550 nm. Sections were imaged with a dry 20× objective (NA 0.70, working distance 0.5 mm), with a pixel size of 1.14 × 1.14 μm2, a z step of 3 μm and a z stack of approximately 25 μm. Fields of view were stitched together to form tiled images using an automated stage and the algorithm of the LAS X software.
Automated cell counting
To perform automatic brain-wide analysis of FOS+ cells, we used the NeuroInfo software (MBF Bioscience). In brief, immunohistochemistry-labelled whole-brain sections were aligned to the Allen Mouse Brain Atlas using the registration tool in the program55. Here brain sections were matched to the most closely corresponding atlas plate. All sections were manually adjusted where necessary, to ensure accurate fit. After the correct atlas plate was identified, all anatomical regions of interest were delineated and measured for size in μm2. Next, FOS+ cells were identified using the cell-detection workflow. Throughout all sections, only cells between 7 μm and 19 μm were counted for size consistency. These parameters were then combined with a deep-learning algorithm to successfully identify FOS+ cells. Counts were then checked for accuracy and adjusted where necessary. Finally, detected cells were mapped to the Allen Mouse Brain Atlas and tallied in the corresponding anatomical brain structure. Normalized counts (cells per μm) were subsequently plotted into GraphPad Prism 10.0 for further analysis.
Network analysis
Cross-correlations between all pairs of regions were calculated in R as previously reported32. Pearson’s correlations were computed in all cases after which networks were created using the igraph package. To be able to interpret the networks, correlations with an absolute value below 0.5 were not included. Both positive and negative correlations were included in the networks.
Metabolic rate and cell activity analysis
To investigate whether FOS and engram activity correlated with metabolic rate, we correlated FOS+ and colabelled eYFP+ (%) with the exact metabolic rate (oxygen consumption, carbon dioxide production and energy expenditure) that was measured 45 min before brain collection.
RNA extraction and qPCR analysis
Collected BAT was defrosted at room temperature and transferred to a 2-ml tube, and stainless-steel beads were added to each individual tube. Next, tissues were homogenized in 1 ml TRIzol reagent in a tissue lyser for 2.5 min at 25 pulses per second. Chloroform (200 µl) was subsequently added to each tube, after which they were inverted and left at room temperature for 3 min. All tubes were then centrifuged at 12,000g for 15 min, the RNA was transferred into a new tube and 500 µl isopropanol was added to precipitate the RNA. All tubes were inverted ten times and left at room temperature for 10 min, before being centrifuged again at 12,000g for 10 min. Supernatants were then discarded and all remaining RNA pellets were washed in 1 ml of 25% RNase-free dH2O and 75% ethanol. After being centrifuged at 12,000g for 5 min, all supernatants were discarded by inverting the tube. RNA pellets were left to dry at room temperature for 20–30 min until they were transparent, after which they were resuspended in 50 µl RNase-free water. Finally, RNA was left on ice for 30 min and subsequently put on a heat block at 55 °C for 15 min. A Nanodrop 2000 UV spectrophotometer (Thermo Fisher Scientific) was used to assess RNA quality and concentration. Next, 20 µl cDNA was synthesized from 2 µg of isolated RNA using a cDNA reverse transcription kit (Biosciences Ltd) in a MiniAmp Thermal Cycler (BD Biosciences). qPCRs were then performed to quantify the relative mRNA expression of genes of interest. Relative mRNA levels were calculated using the ∆∆ cycle threshold (∆∆Ct) method and normalized to corresponding endogenous controls (Ppib). All calculated values were plotted into GraphPad Prism 10 for further analysis.
Stereotactic surgeries
Optic fibre implants
Mice that were used in the optogenetic experiments were implanted with a custom implant containing two optic fibres (200 mm core diameter; Doric Lenses). The optic fibre implant was lowered above the injection site at –1.75 mm dorsoventral. To secure the implant, an even layer of Metabond (C&B Metabond) was applied and left to dry for 15 min. A protective cap was then made from a black polypropylene microcentrifuge tube and secured with dental cement. Animals were then put in a recovery chamber at 29 °C until they were fully recovered from the anaesthesia before being returned to their home cage. Animals were allowed to recover from surgery for approximately 2–3 weeks before behavioural testing.
Viral injections
FOS-tTa transgenic mice were put on a diet containing doxycycline (DOX) 48 h before surgeries. All surgeries were performed when the mice were approximately 7–8 weeks old. Mice were anaesthetized with 500 mg kg–1 avertin and head-fixed on a stereotaxic frame. Bilateral craniotomies were performed using a 0.5-mm diameter drill at –2.00 mm anteroposterior and ±1.35 mm mediolateral. Next, 300 μl AAV9-TRE-ChR2-eYFP was injected on each side using a microsyringe pump (UMP3; WPI) and a Hamilton syringe (701LT; Hamilton) at –2.00 mm anteroposterior, ±1.35 mm mediolateral and –2.00 mm dorsoventral. The injection speed was 60 nl min–1 and the needle was left inside for an additional 10 min after virus delivery to achieve maximum virus spread.
For chemogenetic experiments, TRAP2 mice were injected with 300 μl pAAV-hSyn-DIO-HA-hM4D(Gi)-IRES-mCitrine on each side at –2.00 mm anteroposterior, ±1.35 mm mediolateral and −2.00 mm dorsoventral. The injection speed was 60 nl min–1.
Temperature probe surgery
Before implantation, all temperature probes were sterilized. Mice were anaesthetized and positioned on their back as previously described56. Next, all of the skin around the stomach area was sanitized with 70% ethanol and sterilized saline. A small incision was made using a scalpel in the middle of the abdomen, followed by a 1-cm incision in the peritoneum. A temperature probe was subsequently put in the abdominal cavity. Organs were placed over the probe before sewing the peritoneum and skin with sterile stitches.
Labelling strategies
TRAP2 strategy
The TRAP2 system enables the permanent labelling of neurons that were activated by a given experience57. TRAP2 relies on an immediate early gene locus to drive the expression of iCre recombinase combined with a transgenic Cre-dependent effector. The iCre recombinase can be controlled by tamoxifen. As such, when a neuron is active and tamoxifen or 4-OHT is present, the iCre recombinase can enter the nucleus and result in expression of the effector33. The TRAP2 mice were combined with two different reporter lines. For optogenetic experiments, TRAP2 mice were bred with the Ai32 mice48. For engram cell-counting experiments, TRAP2 mice were crossed with the R26R mice34.
To label the cells that were activated during a cold experience, mice were injected with tamoxifen 5 h after being put in the Promethion cages on CL1. As such, all cells that were active during the 6-h cold exposure were labelled. All mice were returned to their home cages immediately after labelling.
FOS-tTa strategy
Here we used an adeno-associated virus that expresses channelrhodopsin-2 (ChR2)–eYFP under the control of a tetracycline-responsive element (TRE)-containing promoter (AAV9-TRE-ChR2-eYFP). The TRE promoter is only active in cells that contain the tetracycline transactivator (tTa)42. Therefore, we injected the AAV9-TRE-ChR2-eYFP virus into the DG of Fos-tTa transgenic mice. These mice express tTa under the control of a Fos promoter20. Because FOS is activity dependent, the Fos-tTa transgene will only express tTa in cells that are active during a given experience. By combining the transgenic mice with the virus, we were able to express tTa and ChR2–eYFP in cells that were active during a cold experience. To have temporal control over this system, mice were put on a diet that contained DOX. DOX is an antibiotic that binds to tTa and as such prevents the expression of the transgene ChR2-eYFP.
To label cells activated by a cold experience, DOX food was substituted with standard food 36 h before being exposed to a cold experience. Cold memory engrams were identified as the cells that expressed ChR2–eYFP as a result of being active during the cold. To close the labelling window, mice were immediately put back on DOX food after the initial cold experience.
4-OHT
To label cold-sensitive engrams in Trap2;Ai32 and Trap2;R26R transgenic mice, we intraperitoneally injected mice with 4-OHT (50 mg kg–1, Santa Cruz). All 4-OHT was freshly prepared on the labelling day in a mix of sunflower seed oil and castor oil (4:1, Sigma-Aldrich) at 10 mg ml–1.
Engram tagging
To identify the cells that were active during a cold experience, we used Trap2;R26R mice. These mice were chosen because they showed more reliable labelling in the hypothalamus than did other mice. In brief, all mice were injected with tamoxifen 5 h after being put in the Promethion cages on CL1 to label all cells that were active during the 6-h cold exposure. Next, mice were exposed to the rest of the cold memory timeline as described above (two additional training days, a 2-day break and BL2). On the test day, half of the mice were put into context B at 21 °C and half of the mice were kept in their home cage. After exactly 3 h and 45 min all mice were euthanized to enable visualization of the immediate early gene Fos.
Engram counting
All cells were counted using Fiji58. An investigator blinded to treatment counted eYFP+, FOS+ and colabelled cells bilaterally in the DG, LHA, MPO and the LPO. All cell counts were normalized to their respective areas and plotted as cells per μm2.
Optogenetic reactivation of cold memories in metabolic cages
Shining 472-nm blue light on a ChR2-expressing cell causes ChR2 channels to open, resulting in the depolarization of the neuron. This enables us to evoke memory recall in mice with ChR2-labelled cells. Optogenetic activation was performed through a 450-nm laser diode fibre light source (Doric LDFLS 450/080). Mice with optic fibre implants were attached to a patch cord (MFP_200/240/900-0.22_0.3m_FC-ZF1.25(F)) and allowed to habituate for 8 h before being put through the above-described cold memory timeline. Cold-sensitive engrams were labelled on CL1 and optogenetic reactivation was performed one week later in context A. On the reactivation day, mice were exposed to a 15-min optogenetic stimulation (8 mA) session that was divided into 3-min on and 1-min off periods (that is, 3 min on, followed by 1 min off). During the light-on period, mice received blue-light stimulation (20 Hz) with a pulse width of 15 ms for the entire 3-min duration. During the light-off period, the mice received no light stimulation. To enable animals to return to baseline, each 15-min stimulation period was followed by 1 h of no light stimulation. Mice received a total of three rounds of 15-min stimulation periods. Metabolic rates were measured at all times.
Chemogenetic inhibition of cold memories in metabolic cages
To inhibit cold engrams, we used the same labelling timeline as described above. In brief, mice went through two habituation days and a baseline measurement day in context A, at 21 °C. On day 4, contextual cues were added to the chambers and the mice spent a total of 6 h in the altered context at a temperature of 4 °C. To label the neurons that were active during these 6 h of 4 °C cold exposure, mice were injected with 4-OHT 5 h after being put in the Promethion cages. All mice were returned to their home cages immediately after labelling. Mice subsequently went through two additional training days and were then kept in their home cages for a two-day break period. On day 9, mice were put back in the Promethion cages without any of the additional cues at 21 °C to allow for a second baseline measurement (context A). Finally, on day 10, half of the mice were injected with CNO (2 mg kg–1, Tocris) and half of the mice were injected with saline. Mice were kept in their home cage for 30 min after injection and were then put in the Promethion cages (context B, 21 °C).
Statistics
All data were analysed using GraphPad Prism 10. All data are mean ± s.e.m. An α level of 0.05 was used as a criterion for statistical significance and probability levels were quoted for non-significance. All statistical analyses performed are reported with their outcomes in Supplementary Data 1 and 2.
Figure design and visualization
Illustrations were created with BioRender (www.biorender.com).
Inclusion and diversity
We support inclusive, diverse and equitable conduct of research.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.