Mouse lines
All mouse lines were generated by CRISPR–Cas9 at the Genome Engineering Unit (https://crisprmice.eu/) as described previously3,5,7. A conditional Tent5aflox/flox and double-knockout Tent5aflox/floxTent5c−/− have been described previously7. The Cd11c-Cre (B6N.Cg-Tg(Itgax-cre)1-1Reiz/J) mouse strain was purchased from The Jackson Laboratory and crossed with Tent5aflox/flox. The FKBP12F36V-HA-Tent5a mouse line was generated by inserting the sequence of the dTag degron and the 2×HA tag at the N terminus of the endogenous Tent5a. B6CBAF1 zygotes were microinjected with a mixture containing Cas9 mRNA, gRNA targeting Tent5a (AAAAGTATCTCTGATGCATC) and double-stranded repair template (TAGAAGGGGCGGCCGCCTCCAGGTGACACAGACGGGACTCTCGCTTGTGCTTTCCAGATGGGAGTGCAGGTGGAAACCATCTCCCCAGGAGACGGGCGCACCTTCCCCAAGCGCGGCCAGACCTGCGTGGTGCACTACACCGGGATGCTTGAAGATGGAAAGAAAGTTGATTCCTCCCGGGACAGAAACAAGCCCTTTAAGTTTATGCTAGGCAAGCAGGAGGTGATCCGAGGCTGGGAAGAAGGGGTTGCCCAGATGAGTGTGGGTCAGAGAGCCAAACTGACTATATCTCCAGATTATGCCTATGGTGCCACTGGGCACCCAGGCATCATCCCACCACATGCCACTCTCGTCTTCGATGTGGAGCTTCTAAAACTGGAAGGCGGCTACCCCTACGACGTGCCCGACTACGCCGGCTATCCGTATGATGTCCCGGACTATGCAGGCGGACATCAGAGATACTTTTGGTGCGGGGCTGCTCTGCGGGGCGCGGCGCGCGGCTGGGCATTG). Zygotes were transferred to surrogate mothers and pups were screened for the presence of insertion using a 3-primer approach (Fw: GCAGCTCCTGCGAAGTGTG, Rv1: TGCCATGGCAAAGTACCCTTC, Rv2: CCCACACTCATCTGGGCAAC). Potential founders were sequenced by Sanger sequencing. Mice were bred in the animal facility of the Faculty of Biology, University of Warsaw, and maintained under conventional conditions3,5,7 in open polypropylene cages filled with woodchip bedding enriched with nesting material and paper tubes. Mice were fed ad libitum with a standard laboratory diet (Labofeed B, Morawski). Room humidity was maintained at 55 ± 10%, the temperature was kept at 22 °C ± 2 °C, the air was changed at least 15 times per hour and a 12-h–12-h light regime was used (lights on from 06:00 to 18:00). Regular health monitoring was performed at the IDEXX laboratory. Mice of different genotypes were assigned with individual numerical tags in the database and they were used for tissue collection and throughout the subsequent processing as the only identifiers.
Discarded residual vaccine material: mRNA-1273 (Moderna, monovalent or bivalent original/Omicron BA.4-5 spike), BNT162b2 (Pfizer monovalent or bivalent original/Omicron BA.4-5), Nuvaxovid (Novavax, recombinant spike protein, adjuvanted) were used within the manufacturer’s stability guidelines. Because this was not available for purchase at the time and because only residual (otherwise discarded) material could be used, we obtained approval from the Polish Ministry of Health (MMI.454.1.2021.TM).
All animal experiments were approved by the II Local Ethical Committee in Warsaw (approval numbers: WAW2/71/2021, WAW2/129/2021, WAW2/95/2022, WAW2/127/2022 and WAW2/007/2023) and were performed according to Polish law (act number 653 266/15.01.2015) and in agreement with the corresponding European Union directive.
Immunizations
Six-to-fourteen-week-old mice were immunized by intramuscular (Vastus lateralis region) injection with 0.9% NaCl (controls), 1 µg mRNA-1273 (Moderna) or BNT162b2 mRNA (Pfizer) or 50 µl Nuvaxovid vaccine (Novavax) at 40 ng µl−1 protein concentration. Tissues were collected at set time points for subsequent analyses. Mice of both sexes were used, with equal or similar ratios of both sexes in directly compared groups. The sex of the mice was not considered in the immunization experiments, only the genotype.
No formal sample-size calculations were performed a priori. Sample sizes were determined on the basis of established practices in similar published studies and the 3Rs principles (replacement, reduction and refinement) for ethical animal research. We used the minimum numbers necessary to achieve statistical significance while maintaining scientific rigour, on the basis of our previous experience and comparable studies. Details on sample sizes are provided for each experiment. Mice with different genotypes could not be distinguished by the experimenter (except for Tent5a−/− mice, which have a distinguishable phenotype). However, vaccination is a routine procedure and therefore knowledge about the genotype of mice at the time of vaccination was highly unlikely to affect the results of immunization.
ELISA
Concentrations of spike protein receptor-binding domain, anti-spike IgG and total IgG in mice serum were measured according to the manufacturer’s recommendations with commercially available ELISA kits: Invitrogen (EH492RB; 1222062723, 1222110123 and 1222110723), Krishgen Biosystems (KBVH015-14; MACOVSPQ1023, MACOVSPQ0223 and MCOV2SS0822) and Eagle Biosciences (OGG11-K01; 25Q1 and 26), respectively. The level of OVA was measured using an ELISA kit from MyBioSource (MBS2000240; L240916286). Sample dilution was determined experimentally to fit within the standard curve of each ELISA. Owing to intervals between immunizations, different versions and lots of both vaccines and ELISA kits were used, depending on availability. Therefore, ELISA results should be interpreted as semi-quantitative between experiments. A single experiment was always performed simultaneously for all conditions studied, and can therefore be interpreted as quantitative, allowing direct comparisons between experimental conditions.
Sorting infiltrating macrophages and DCs from muscles
Muscle dissociation and isolation of immune cells
Muscles from anterior thigh were isolated 6 h, 12 h, 24 h or 48 h after vaccine injection, finely minced using surgical scissors and resuspended in 2 ml of serum-free RPMI supplemented with 1,000 U ml−1 collagenase from Clostridium histolyticum (Sigma-Aldrich, C9407), 250 µg ml−1 DNAse I (Sigma-Aldrich, 10104159001) and 8 µg ml−1 dispase I (Sigma-Aldrich, D4693). Samples were incubated for 30 min at 37 °C with 140-rpm agitation. Digested muscle suspensions were put on ice and processed at 4 °C from then on. Ten millilitres of ice-cold FACS buffer (0.2% BSA in PBS) was added to muscle suspensions, followed by straining with a 70-µm nylon strainer (Greiner, 542070). With the strainer on, the suspension was centrifuged at 300g for 7 min. The crude pellet was treated twice with ACK (150 mM ammonium chloride, 1 mM potassium bicarbonate and 0.1 mM disodium EDTA, pH 7.2) for 2 min on ice, followed by washing with 10 ml FACS buffer; each time, centrifugation was performed at 300g for 7 min. The crude pellet was then resuspended in 1 ml FACS buffer and strained through the 30-μm cap strainer of a FACS tube. With caps on, the samples were centrifuged at 800g for 3 min at 4 °C and the supernatant was discarded, followed by resuspension in 2 ml FACS buffer. Equal volumes of samples were mixed thoroughly with 33% Percoll (GE17-0891-01) in PBS and spun at 800g for 30 min at 4 °C. The supernatant was discarded and the cell pellet was subjected to staining.
Cell staining and sorting
Cells were washed once with FACS buffer and incubated with Fc block anti-CD16/32 (clone 2.4G2) (BD Biosciences, 553142) for 5 min on ice, washed with FACS buffer and incubated with anti-CD45–PerCP–Cy5.5, anti-CD11b–BV786, anti-F4/80–PE–Cy7, anti-I-A/I-E–BV605, anti-CD11c–PE and anti-CD64–APC–Fire750 for 40 min in 4 °C, protected from light (detailed information on antibodies and dilutions is provided in Supplementary Table 11). After staining, cells were washed with FACS buffer and then stained with the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Thermo Fisher Scientific, L34964) for 15 min at 4 °C, protected from light. Cell sorting was performed in a CytoFlex SRT cell sorter (Beckman Coulter) operated by CytExpert SRT v.1.1 software. A restrictive gating strategy was used to sort M0 macrophages and DCs (Supplementary Fig. 5). The gating strategy was based on gating live singlets, next cells identified as CD45+, CD11c+ and I-A/I-E (MHCII)+, with the exclusion of cells that were double positive for CD64 and F4/80, were sorted as DCs. Live singlets identified as CD45+, CD11c+, I-A/I-E (MHCII)low/−, F4/80+ and CD64+ were sorted as M0 macrophages. For post-sort analysis, cells were sorted to 100 µl of PBS. Post-sort analysis was performed on the first sample, to verify the purity of sorted populations. For RNA isolation, cells were sorted directly into 100 μl or 250 µl of extraction buffer (Applied Biosystems, KIT0204), depending on the cell amount, followed by centrifugation at 14,000g for 5 min and freezing until further processing.
Cell cultures
A549 and HEK293 Flp-In T-REx cell lines and their derivatives were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS (Gibco) and penicillin–streptomycin (Sigma-Aldrich) at 37 °C in a 5% CO2 atmosphere until 80% confluency. To produce the HEK293 Flp-In T-REx (R78007, Thermo Fisher Scientific) cell line with conditional knockdown of CNOT1, we used a strategy previously described by us29. A tri-miRNA construct (listed below; stem–loop sequences ATGGAAGAGCTTGGATTTGAT, CTCCCTCAATTCGCCAACTTA and AGGACTTGAAGGCCTTGTCAA) was designed, and cloned at the BspTI (AflII) + NotI restriction sites in a pKK-RNAi vector (pKK-BI16 nucCherry EGFP-TEV). A total of 1 × 106 HEK293 Flp-In T-REx cells, grown in DMEM high-glucose medium (Gibco) supplemented with 10% FBS on six-well plates, were transfected with 300 ng of the CNOT1-bearing construct mixed with 1 μg of pOG44 plasmid and 250 μl of Opti-MEM medium, supplemented with 2 µl TransIT-2020 transfection reagent (Mirus, MIR5400). The cells were seeded in a 60-mm plate and cultured in DMEM high-glucose medium supplemented with 40 µg ml−1 hygromycin and 8 µg ml−1 blasticidin for the first six to seven days. Cells were grown until single colonies appeared four weeks later. A control of cells that were not transfected with the pKK-BI16 plasmid was included to ensure the specific selection of the cell line. Expression of exogenous miRNA genes was induced by the addition of doxycycline (Thermo Fisher Scientific) at a final concentration of 100 ng ml−1. Cell enumeration was performed by crystal violet (Sigma, C3886) staining described by the laboratory of X. Chen (UCSF). In brief, cell medium was aspirated (or collected for subsequent ELISAs if necessary) and cells were washed with 1 ml PBS. Staining solution (0.25% crystal violet in 20% methanol) was then added to each well and cells were incubated for 10 min at room temperature. The staining solution was then removed, and the cells were rinsed 6 times with 2 ml PBS, after which the remaining solution was evaporated, and the plates were scanned.
mBMDM cultures
The primary mBMDM cultures were established from the bone-marrow monocytes isolated from Tent5aflox/floxTent5c−/−, FKBP12F36V-HA-Tent5a and wild-type mice. Young adult mice (12–25 weeks) of both sexes were euthanized by cervical dislocation. The femurs and tibias were isolated and bone marrow was collected using a centrifugation-based protocol24,30. Material was mixed from several individual mice (siblings of the same sex) to obtain sufficient cells for subsequent analyses. The sex of the mice used as a source of bone marrow cells was not considered in further analyses. Bone-marrow cells were plated in IMDM medium (Thermo Fisher Scientific, 21980065) supplemented with 10% FBS (Gibco), 100 U ml−1 penicillin and 0.1 mg ml−1 streptomycin solution (Sigma-Aldrich) and 10 ng ml−1 macrophage colony-stimulating factor (M-CSF; PeproTech, 315-02), and cultured at 37 °C in 5% CO2. For conditional Tent5a gene targeting, mBMDMs were transduced on the eighth day after isolation with lentivirus carrying Cre recombinase (pCAG-Cre-IRES2-GFP). Lentivirus production, cell transduction and genotyping were performed as described previously3,7. Cells were used for experiments on the fourteenth day after isolation.
To induce depletion of TENT5A, 0.7 × 106 mBMDMs obtained from wild-type and FKBP12F36V-HA-Tent5a mice were seeded in six-well plates and 24 h later, degron was induced by the addition of 1 µM dTAGv-1 or dimethyl sulfoxide (DMSO) as a control31. The vaccine was added to cells at the same time and cells were collected in time courses at set time points.
Analysis of translation dynamics in mBMDMs
mBMDMs were reversely transfected with IVT mRNA (with uridines or mΨ) encoding high-RSCU EGFP (ref. 32) using MessengerMAX (Invitrogen, LMRNA001) according to the manufacturer’s instructions in five technical replicates. In brief, the mRNA used for transfection was diluted in Opti-MEM (Gibco, 31985047) to obtain 40 ng of mRNA in 10 µl of Opti-MEM, and MessengerMAX (Invitrogen, LMRNA001) was diluted in Opti-MEM equivalent to 0.1 µl of reagent in 10 µl of Opti-MEM per well. Diluted mRNA was mixed with diluted transfection reagent on 96-well plates, left for 40 min and then transferred to 384-well plates (Perkin Elmer, 6057300). mBMDMs were counted in a Countess 3L counter (Invitrogen, AMQAF2000) using dedicated chamber slides (Invitrogen, C10283). Finally, cells were diluted to 5,000 cells per well and supplemented with dTAG-V1 (Bio-Techne, 7374/5) to a final concentration of 2 µM. Control cells were seeded without degron inductor. Cells were then seeded at a volume of 100 µl onto prepared plates with transfection mixes using the Multipette E3x (Eppendorf, 4987000029) and Combitip 5 ml Biopur (Eppendorf, 0030089669). The imaging was started 4 h after transfection in Opera Phenix from Perkin Elmer using three channels: 488 nm (time: 50 ms, power: 20%, height: −8.0 µm); bright-field (time: 20 ms, power: 20%, height: −0.0 µm); digital phase contrast (time: 20 ms, power: 20 %, height: −1.0 µm); water objective 20× non-confocal; binning 2 × 2. Live images were taken every 2 h for 72 h.
Isolation of human CD14+ cells
Buffy coats were obtained commercially from the Regional Blood Centre in Warsaw. All donors were 18–45-year-old healthy men. Peripheral blood mononuclear cells (PBMCs) were isolated within 1–2 h after donation by density-gradient centrifugation using Lymphoprep (STEMCELL Technologies). Next, CD14+ PBMCs (monocytes) were isolated immunomagnetically with anti-human CD14 antibody-coated microbeads using LS columns (all from Miltenyi Biotec), strictly following the manufacturer’s protocol. CD14+ cells were counted, and viability was checked in 0.1% trypan blue solution (Sigma-Aldrich) and used for subsequent hMDM differentiation. Each isolation resulted in a viability of over 95%.
In vitro culture of hMDMs
CD14+ PBMCs were seeded in a 10-cm tissue-culture-treated Petri dish at a density of 1 × 106–2 × 106 cells per ml in 10 ml of RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% heat-inactivated FBS (HyClone), 2 mM l-glutamine (Sigma-Aldrich) 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (both Sigma-Aldrich), 1% (v/v) MEM non-essential amino acids solution (Thermo Fisher Scientific) and 1 mM sodium pyruvate (Sigma-Aldrich) supplemented with 50 ng ml−1 of recombinant human M-CSF (for macrophage differentiation). Adherent cells collected from the M-CSF-supplemented culture were checked for markers of macrophages. hMDMs were stained with Zombie UV or Zombie Aqua Fixable Viability Kit (BioLegend) and antibodies detecting membrane markers CD14, CD83, CD86, CD163, CD206 and HLA-DR (listed in Supplementary Table 11). The antibody panel was optimized using isotype and fluorescence minus one (FMO) controls and a built-in compensation algorithm. Flow cytometry was performed using a Fortessa X-20 Analyzer (BD Biosciences) operated by FACSDiva 8.3 software. FlowJo v.10.6.1 software (BD Biosciences) was used for data analysis.
Administration of mRNA LNPs and mRNA to in vitro-cultured cells
The 0.5 × 106–1 × 106 cells were seeded the day before on a six-well plate in medium as described above. The required amount of vaccine mRNA in original LNP formulation was diluted in 150 µl of Opti-MEM medium (Thermo Fisher Scientific) at room temperature, and after 10 min was added to the cells in a drop-wise manner and gently mixed. Cells were collected at time points specified for the individual experiments.
In the dose-dependent experiment, cells were transfected with varying volumes of original LNPs, with the quantity of RNA required (0.2 µg, 1 µg, 2 µg, 5 µg or 10 µg) diluted in OPTI-MEM medium. For the control cells (those not treated with the specified agent), an appropriate volume of OPTI-MEM was added. All transfections of purified vaccine mRNAs were performed with Lipofectamine MessengerMAX (Thermo Fisher Scientific), according to the manufacturer’s instructions. Cells were collected at time points specified for the individual experiments for subsequent analyses.
Cell fractionation
Macrophages were fractionated using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific, 78840), according to the manufacturer’s instructions. All buffers used were supplemented with RiboLock (Thermo Fisher Scientific, EO0382) to a final concentration of 1 U µl−1. The collected fractions for RNA isolation were immediately combined with TRIzol LS, incubated for five minutes at room temperature and subsequently frozen at –80 °C. For western blot analyses, samples were frozen in liquid nitrogen.
Gene silencing by siRNA
siRNA-mediated knockdowns in hMDMs were performed using validated stealth siRNAs: HSS124645, HSS124646 and HSS183139 (all for TENT5A). Knockdowns in mBMDMs were performed using validated stealth siRNAs: MSS279478 (Tent3a; also known as Tut4 and Zcchc11), MSS279479 (Tent3a), MSS210722 (Tent3b; also known as Tut7 and Zcchc6), MSS210723 (Tent3b), MSS200078 (Tent2; also known as Papd4), MSS200079 (Tent2), MSS209990 (Tent4a; also known as Papd7), MSS209991 (Tent4a), MSS277931 (Tent4b; also known as Papd5), MSS277932 (Tent4b), MSS220103 (Fndc3a) and MSS291871 (Fndc3b).
Transfections were performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions, as previously described24,33. Recommended negative controls (Invitrogen, 12935100) were used.
General molecular biology techniques
Western blots
An equal number of cells were lysed in PBS supplemented with 0.1% NP40, protease inhibitors and viscolase (final concentration 0.1 U ml−1; A&A Biotechnology, 1010-100) for 30 min at 37 °C with shaking at 1,200 rpm. Then 3× SDS sample buffer (187.5 mM Tris-HCl pH 6.8, 6% SDS, 150 mM DTT, 0.02% bromophenol blue, 30% glycerol and 3% 2-mercaptoethanol) was added and samples were boiled for 10 min. Samples were resolved on 12–15% SDS–PAGE gels and then proteins were wet-transferred to Protran nitrocellulose membranes (GE Healthcare) at 400 mA at 4 °C for 1.5 h in 1× transfer buffer (25 mM Tris base, 192 mM glycine and 20% methanol (v/v). Next, the proteins were visualized by staining with 0.3% w/v Ponceau S in 3% v/v acetic acid and digitalized. Membranes were blocked by incubation in 5% milk in TBST buffer for 1 h followed by overnight incubation at 4 °C with specific primary antibodies (listed in Supplementary Table 11) diluted 1:2,500 (spike protein, CD80, GRP94, HA, SSR1, PERK, calreticulin and PDI) or 1:5,000 (α-tubulin, β-actin, actinin, eIF2a and GAPDH). Membranes were washed three times in TBST buffer for 10 min each and incubated with HRP-conjugated secondary antibodies (anti-mouse (Millipore, 401215) diluted 1:5,000 and anti-rabbit (Millipore, 401393) diluted 1:5,000) for 2 h at room temperature. Membranes were washed three times in TBST buffer and proteins were visualized using X-ray films or ChemiDoc (BioRad) or iBright (Thermo Fisher Scientific) imaging systems. Blots were quantified using Multi Gauge v.3.0 (Fujifilm Life Sciences). Unprocessed scans are shown as source data.
RNA isolation
Total RNA was isolated from cells or vaccine samples with TRIzol reagent or TRIzol LS reagent (both from Thermo Fisher Scientific), respectively, according to the manufacturer’s instructions, dissolved in nuclease-free water and stored at −20 °C (short term) or −80 °C (long term). RNA from frozen muscles and lymph nodes was isolated by tissue homogenization in TRI reagent (Sigma, T9424) pre-heated to 60 °C, using Omni Tissue Homogenizer equipped with a 7 × 115-mm Saw Tooth (Fine) Generator Probe. Homogenous mixtures were further processed according to the manufacturer’s instructions. For qRT–PCR, RNA-seq and DRS library preparation, the RNA was treated with TURBO DNase (Thermo Fisher Scientific, AM1907; or Invitrogen, AM2238). To assess the integrity of isolates, each RNA sample after DNAse treatment was analysed with the Agilent 2200 TapeStation system, using Agilent High Senitivity RNA ScreenTape (Agilent, 5067-5579).
RNA from cells acquired by fluorescence-activated cell sorting (FACS) was isolated using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, KIT0204) following guidelines for RNA isolation from cell pellets with on-column DNAse treatment (Qiagen, 79256), with some modifications. For qRT–PCR analyses, we sorted 1,000 cells of each type, from separate mice, directly to 100 µl extraction buffer. In the case of cDNA sequencing, we sorted 20,000 cells of each type, pooled from three mice, directly to 250 µl extraction buffer. During the precipitation step, we used a 1:1 (v:v) 70% ethanol:extraction buffer ratio. RNA was eluted in 15 µl elution buffer in every case.
qRT–PCR for mRNA-1273 quantification and relative gene expression
cDNA was synthesized using 500 ng of DNAsed total RNA as a template with SuperScript III Reverse Transcriptase (Invitrogen, 18080093) according to the manufacturer’s instructions. The final cDNA concentration was kept at 2.5 ng µl−1 (converted from total RNA). In the case of RNA from FACS-sorted cells, 11 µl of the entire eluate was used for cDNA synthesis.
To determine the concentration of Moderna’s mRNA-1273 in mouse tissues or FACS-sorted cells, a custom-made TaqMan probe together with TaqMan Gene Expression Master Mix (Applied Biosystems, 4369016) was used and normalized with a pre-designed gene-expression assay for β-actin (Applied Biosystems, Mm01205647_g1). The reaction mix was contained in a total volume of 10 µl with 1× concentration of master mix and gene-expression assay; 5 ng cDNA was used per reaction (converted from total RNA). In the case of FACS-sorted cells, 2 µl of 1.5× diluted cDNA was used, which roughly translates to 60 cells per 384-well plate, assuming 100% recovery of material during RNA isolation and 100% efficiency of cDNA synthesis. A thermal cycling program for TaqMan Gene Expression Master Mix was used as instructed by the manufacturer.
To estimate the relative expression of Tent5a and Tent5c at injection sites or in FACS-sorted cells, and the levels of mRNA-1273 in HEK293T and A549 cells with varying amounts of vaccine, we used Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, 11733046) following the general protocol for ABI instruments recommended by the manufacturer.
For all qRT–PCR analyses, the QuantStudio 5 Real-Time PCR System, 384-well (Applied Biosystems, A28140) was used.
Custom TaqMan probe design and absolute quantification of vaccines
The primer blast algorithm was used to find a unique amplicon for mRNA-1273 in the mouse transcriptome (Refseq mRNA). Within the amplified region, a 15-mer of optimal GC content (53.3%) was selected as the target site for probe hybridization. We selected FAM dye and MGB-NFQ quencher as 5′ and 3′ modifications for the probe. The probe was synthesized by the Thermo Fisher Scientific Custom TaqMan probes service. To test the specificity and sensitivity of the designed assay, we performed a serial dilution experiment in which we spiked 500 ng of DNAsed mouse total RNA with 50 ng of DNAsed mRNA-1273 and performed cDNA synthesis. Then we performed 10× serial dilutions of that mix with unspiked mouse cDNA of the same concentration. qRT–PCR analysis showed that the designed assay is specific and can detect up to 10 ag of mRNA-1273 per ng of total RNA, which is equivalent to roughly 25 molecules. Similarly, we prepared a standard curve with 10× serial dilutions of mRNA-1273 to estimate the absolute concentration of vaccine in FACS-acquired cells. The concentration of vaccine before cDNA synthesis was determined with the Agilent 2200 TapeStation system using Agilent High Sensitivity RNA tape. Five nanograms of vaccine was added to 500 ng yeast total RNA and cDNA was synthesized; the final sample volume was 200 µl. Next, we performed eight serial dilutions (10×) using yeast cDNA of 2.5 ng µl−1 concentration. Recorded Ct values for each concentration served as data points for plotting an exponential curve, and we used this equation to convert Ct values to molecules of mRNA-1273, assuming its molecular mass is 1321.81 kDa. Next, the calculated number of molecules was normalized to the number of cells in a 384-well plate (translated from the % of eluate used for cDNA synthesis, assuming 100% recovery of cells during sorting and RNA isolation as well as cDNA synthesis efficiency).
qRT–PCR for ER stress assessment
The relative expression of genes associated with ER stress, we assessed CHOP (also known as DDIT3), GRP94 (HSP90B1), PERK (EIF2AK3) and XBP1 mRNA splicing in mRNA-1273-transfected hMDMs (wild-type and TENT5A knockdown) using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, 11733046), following the general protocol for ABI instruments recommended by the manufacturer. Gene expression for each sample was normalized to ACTB.
Primers and probe sequences are listed in Supplementary Table 12.
3′-RACE sequencing
To examine the 3′-UTR and terminal sequence of the mRNA-1273, RNA was freshly isolated from the vaccine sample vial. Then, 1 µg of vaccine mRNA was ligated to 20 pmol RA3_7N adaptor: 5rApp/CTGACNNNNNNNTGGAATTCTCGGGTGCCAAGG/3ddC with 10 U of T4 KQ227 RNA ligase 1 (NEB, M0204S) in the presence of 20 U RNase OUT (Thermo Fisher Scientific, 10777019), 1× T4 RNA Ligase Reaction Buffer (NEB, M0204S), 1 mM ATP and 20% PEG8000 in a total reaction volume of 20 µl at 25 °C for 4 h. Ligase was inactivated at 65 °C for 20 min. The ligation product was purified with a 0.8× ratio of KAPA Pure Beads and eluted with 15 µl RNase-free water, according to the manufacturer’s protocol. The cleaned ligation product was subjected to reverse transcription with 40 pmol of Illumina index adapter: 5′-CAAGCAGAAGACGGCATACGAGATATCAGTGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′ with SuperScript III (Thermo Fisher Scientific, 18080093), 1× First Strand Buffer (Thermo Fisher Scientific, 18080093), 0.25 mM dNTP mix, 5 mM DTT and 20 U RNAse OUT (Thermo Fisher Scientific, 10777019). The reaction mix was incubated at 45 °C for 1 h and 70 °C for 20 min in a thermocycler. The reverse transcription product was cleaned with a 1× ratio of KAPA Pure Beads and eluted with 19 µl RNase-free water. Prepared cDNA was diluted in a 1:3 proportion. In the library amplification step, 0.5 µl of cDNA was mixed with 0.4 pmol of gene-specific starter 5′-AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGACGATCAGAAGGAGATCGATCGGCTG-3′, 0.2 pmol of RP universal starter 5′-CAAGCAGAAGACGGCATACGAGAT-3′, 0.25 mM dNTP, Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, F530S), 1× Phusion HF Buffer (Thermo Fisher Scientific, F530S) and 3% DMSO. For amplification, a standard Phusion program was used at 65 °C for annealing and 25 cycles. The 50 µl of PCR product was separated in 2.5% agarose gel. A band of 450 bp was cut out from the gel and purified with Gel-OUT (A&A Biotechnology, 023-50) according to the kit protocol. The library was cleaned twice with a 1.0× ratio of KAPA Pure Beads. TapeStation analysis of the sample was performed as a quality control. The library was sequenced on an Illumina NovaSeq 6000 sequencer.
Analysis of data from 3′-RACE sequencing
RA37_N adapter sequence was trimmed from the R2 read (containing poly(A) tail) with cutadapt34 (options -g CCTTGGCACCCGAGAATTCCANNNNNNNGTCAG –discard-untrimmed). Then only reads containing poly(A) tail were identified with cutadapt (options -a TTTTTT –discard-untrimmed –fasta). Obtained sequences were reverse complemented using fastx_reverse_complement from the fastx toolkit (0.0.13) and loaded into R with the BioStrings package. To get rid of unwanted trimming artefacts, sequences with length between 0 and 10 were chosen, four A letters (representing the poly(A) tail) were pasted before each sequence for visualization purposes and a sequence logo representing the nucleotide composition of the 3′ end of mRNA-1273 was produced with the ggseqlogo package35.
Analysis of translation in vitro
For in vitro translation, the Retic Lysate IVT Kit (Invitrogen, AM1200M) was used. Reactions were assembled using the default buffer suggested by the producer. The translation mix (–met) was supplemented with unlabelled methionine to a final concentration of 50 μM. Either 1 μg or 3 μg of purified capped RNA was used as template in each reaction. The reaction was run for 180 min. Spike protein was detected in the reaction mixture using ELISA.
Preparation of standards with predefined poly(A) lengths
Spike-in RNAs were IVT from a set of double-stranded DNA fragments. Templates for transcription were prepared in two consecutive PCR reactions. First, a desired fragment of Renilla luciferase from pRL5Box plasmid was amplified using RLuc_F1/R1-specific primers containing an overhang common for all primers used in the second round of PCR (Supplementary Table 12). PCR products were verified through gel electrophoresis. Correct amplicons were used as templates in the second PCR reaction with RLuc_T7_F2 primer, hybridizing to the overhang sequence from RLuc_F1 and containing the T7 promoter, and backward primer RLuc_Ax_R2, hybridizing to the overhang sequence from RLuc_R1 oligo and introducing a poly(A) tail of a defined length (from 10 to 120 A). The resulting PCR products were assessed and purified by gel electrophoresis. The in vitro transcription reaction was performed at 37 °C for 1.5 h in a 50-µl reaction volume containing 600 pmol T7 template, 10 µl of 5× transcription buffer (200 mM Tris-HCl, 30 mM MgCl2, 10 mM spermidine and 50 mM NaCl), 5 µl of rNTP mix (20 mM each), 5 µl of 100 mM DTT, 0.5 µl of 1% Triton X-100, 80 U ribonuclease inhibitor and 100 U T7 RNA polymerase. Then, DNA template was removed with TURBO DNase (Ambion) for the next 15 min. Spike-in RNAs were phenol–chloroform extracted, precipitated, visually assessed by denaturing electrophoresis, purified on RNA purification beads and used as controls in DRS runs. In each DRS run, a mixture of spike-ins representing RNAs with poly(A) tail of defined lengths (10 A, 15 A, 30 A, 45 A, 60 A, 90 A and 120 A) was included.
EGFP and reporter mRNA synthesis
The EGFP and reporter mRNAs were IVT from plasmid template and double-stranded DNA fragments, respectively. Template for transcription of EGFP mRNA was prepared as follows: the high-RSCU EGFP32 CDS was purchased from Invitrogen and cloned into the plasmid vector pJAZZ (Lucigen) using BigEasy-TSA competent bacteria (Lucigen), providing it with the 5′-UTR and 3′-UTR of the mRNA-1273, a poly(A) tail of 90 A and the T7 promoter Φ6.5 (TAATACGACTCACTATAGGG). The plasmid was digested at the poly(A) tail terminus (PaqCI, NEB) and was verified to be completely digested through agarose gel electrophoresis. DNA template was purified by buffered phenol–chloroform extraction and ethanol precipitation. Templates for transcription of reporter mRNAs were prepared in a PCR reaction (Supplementary Note 6 and Extended Data Fig. 8a,b) as follows: full-length CDSs of Plasmodium falciparum circumsporozoite (CS) protein (PfCSP_3D7; NCBI reference sequence: XP_001351122.1; Extended Data Fig. 7h), OVA (sequence available at https://www.trilinkbiotech.com/) and Zika virus protein E (ZIKVE; NCBI reference sequence: YP_002790881.1) with UTRs from either mRNA-1273 or BNT162b2 were synthesized using the GeneArt service (Thermo Fisher Scientific). For OVA and ZIKVE constructs, an N-terminal ER signal peptide sequence was added (MFVFLVLLPLVSSQCV). All CDSs were optimized for expression in mouse cells using software from Benchling (https://www.benchling.com/). DNAs were PCR-amplified using primers complementary to UTRs with overhangs introducing the RNA T7 promoter Φ6.5 (TAATACGACTCACTATAGGG) at the 5′ end and a poly(A) tail from either mRNA-1273 or Pfizer BNT162b2 and either with or without a TCTAG pentamer at the 3′ end (Supplementary Table 12). In the case of OVA DNAs, an additional set was PCR-amplified as described above, but with primers with a point mutation in the T7 promoter sequence, allowing for efficient cotranscriptional RNA capping (TAATACGACTCACTATAAGG). PCR products were verified through agarose gel electrophoresis and purified with KAPA Pure Beads (KAPA Biosystems).
The composition of the in vitro transcription reaction was: 40–80 ng µl−1 of DNA template, transcription buffer (40 mM Tris-HCl, 26 mM MgCl2, 10 mM NaCl, 2 mM spermidine and 10 mM DTT), T7 RNA polymerase36 (0.04 µg µl−1, in-house prepared), 5 mM ATP/CTP (Thermo Fisher Scientific), 5 mM N1meΨTP (Advent Bio) or UTP (Thermo Fisher Scientific) and 4 mM GTP (in the case of EGFP and OVA reporter mRNAs intended for cotranscriptional capping; Thermo Fisher Scientific) or 5 mM GTP (in the case of all other reporter mRNAs; Thermo Fisher Scientific). The reaction mixtures were supplemented with 1 U µl−1 RiboLock RNase inhibitor (Thermo Fisher Scientific), 0.002 U µl−1 inorganic pyrophosphatase (Thermo Fisher Scientific) and, in the case of EGFP and OVA reporter mRNAs intended for cotranscriptional capping, 10 mM of the trinucleotide Cap1 analogue m7GpppAmpG37 (in-house prepared). The reactions were performed for 120 min at 37 °C. In vitro transcription products were verified through denaturing agarose gel electrophoresis and FPLC-purified using a 0.2 ml GoPure column with POROS Oligo (dT)25 Affinity Resin (Thermo Fisher Scientific) according to the manufacturer’s protocol. The concentration of purified mRNAs was determined using UV absorbance measurements. The purity and integrity of the RNA preparations were determined by the automated electrophoresis system (TapeStation 2200, Agilent Technologies; Extended Data Fig. 8c). The reporter RNAs (except EGFP and cotranscriptionally capped OVA) were then provided with Cap1 using the Vaccinia Capping System (NEB) and Cap 2′-O-methyltransferase (NEB) according to the manufacturer’s protocol with the replacement of the manufacturer’s VCE with the in-house prepared one38. Capped mRNAs were purified with KAPA Pure Beads (KAPA Biosystems). The purity and integrity of the mRNA preparations were determined by automated electrophoresis (TapeStation 2200, Agilent Technologies; Extended Data Fig. 7d).
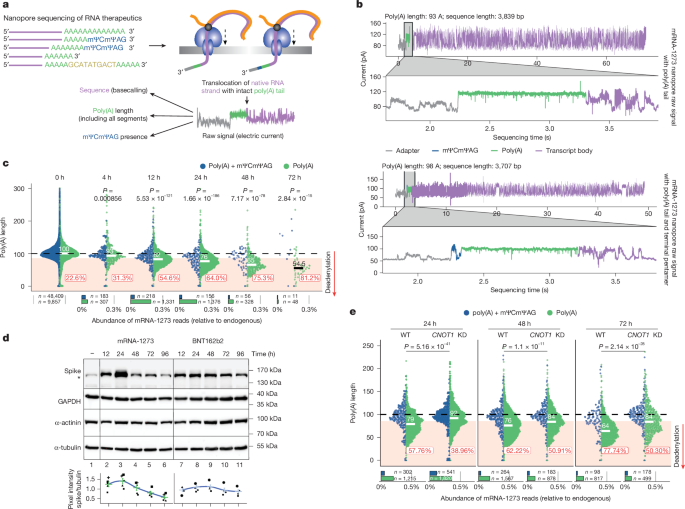
Inosine tailing and nanopore sequencing
Two micrograms of RNA isolated from a sample of mRNA-1273 vaccine was denatured in the presence of 20 U RNAse OUT (10777019, Thermo Fisher Scientific) at 65 °C for 3 min, and immediately placed on ice. An inosine (I)-tailing reaction was performed with 0.5 mM ITP (inosine triphosphate), 1× NEB 2.0 buffer (B7002, NEB) and 2 U poly(U) polymerase (M0337S, NEB) at 37 °C for 45 min, and was terminated by snap-freezing in liquid nitrogen. I-tailed RNA was cleaned twice on KAPA Pure Beads (7983298001, Roche) in a 1× ratio and used for DRS library preparation. I-tailed samples require ligation of a special adaptor RTA_C10, which contains 10 cytosines at the 3′ end: 5′-GAGGCGAGCGGTCAATTTTCCTA AGAGCAAGAAGAAGCCCCCCCCCCCC-3′. To make the I-tailing-specific RTA adaptor, we followed the ONT Direct RNA Sequencing—Sequence-Specific protocol; 0.5 µg of I-tailed mRNA-1273 was taken to the first ligation step of DRS library preparation. Library preparation was performed as described in the Direct RNA Sequencing (ONT, SQK-RNA002) protocol.
Nanopore sequencing
DRS
DRS was performed as described previously3. For raw vaccine isolate, up to 0.5 µg of RNA was used for library preparation. For RNA isolates obtained from cell cultures or mouse tissues, 3.5–5 µg of total mRNA was mixed with 50–200 ng oligo-(dT)25-enriched mRNA from Saccharomyces cerevisiae and standards with predefined poly(A) lengths, and was processed with a Direct RNA Sequencing Kit (SQK-RNA002, ONT) according to the manufacturer’s instructions.
cDNA sequencing
For cDNA sequencing, 400 ng of total RNA was used for library preparation using the cDNA-PCR Sequencing Kit (SQK-PCS111) or PCR-cDNA Barcoding Kit (SQK-PCB111.24 or SQK-PCB114.24) according to the manufacturer’s instructions. Libraries were amplified with 14 cycles of PCR reaction, and the DNA concentration was measured using the Qubit 1× dsDNA High Sensitivity Kit (Invitrogen, Q33231). Then, if multiplexed, they were diluted and mixed equally to reach a final concentration of 60–100 fmol.
cDNA libraries for sequencing with an additional amplification step of mRNA-1273 (applicable for samples with an expected low prevalence of vaccine mRNA, such as cells sorted from vaccine injection sites) were prepared using the standard SQK-PCS111 protocol with the following modification. After synthesis of the first cDNA strand, samples from the reverse transcription reaction were pre-amplified in a 12-cycle PCR reaction with RTP and SSPII_Mod_2 (specific for mRNA-1273, with UMI sequence TTTCTGTTGGTGCTGATATTGCTTTVVVVTTVVVVTTVVVVTTVVVVTTTCCACCGACAACACCTTCGTGAGCGG) primers using the LongAmp Hot Start Taq Master Mix. The reaction products were then purified on Kappa beads, washed with SFB buffer, eluted with H2O and used for the subsequent PCR reaction with barcoded primers, performed according to the SQK-PCB111-24 protocol (14 cycles, 5 min amplification). Finally, cDNA concentrations were measured with Qubit and 12 libraries containing a total of around 60 ng cDNA were used for ligation with the RAPT adapter and sequenced.
Sequencing was performed using R9.4 (for DRS and v.11 DNA chemistry) or R10.4 (for v.14 DNA chemistry) flow cells on a MinION device (ONT) and with the Flow Cell Priming Kit (EXP-FLP002). Raw data were basecalled using Guppy (ONT) in the case of DRS, or dorado (ONT) in the case of cDNA sequencing. Raw sequencing data (fast5 or pod5 files) were deposited at the European Nucleotide Archive (ENA, project PRJEB53190; Supplementary Table 13).
Determining poly(A) lengths from DRS
Basecalled nanopore reads were mapped to the respective transcriptome references (Gencode 26 or Gencode 38 for mouse and human samples, respectively) using Minimap2 2.17 with options -k 14 -ax map-ont –secondary=no, and processed with Samtools 1.9 to filter out supplementary alignments and read mapping to reverse strand (Samtools view -b -F 2320). The poly(A) tail lengths for each read were estimated using the nanopolish 0.13.2 polya function23.
For the analysis of mRNA-1273-originating reads, the nanopolish polya algorithm was modified (Supplementary Note 2) to (1) include unmapped reads enabling the analysis of poly(A) lengths for sDTW-identified reads (Supplementary Note 1) and (2) detect mΨCmΨAG at the 3′ end of the poly(A) tail and report its presence in the output.
For the analysis of BNT162b2-originating reads, the nanopolish polya algorithm was modified analogously (Supplementary Note 3) to (1) include unmapped reads enabling the analysis of poly(A) lengths for sDTW-identified reads and (2) detect two poly(A) segments interleaved with a 10-nt linker and report their lengths in the output.
Determining poly(A) lengths from cDNA sequencing
For cDNA barcoded libraries (SQK-PCB111-24), raw sequencing data were basecalled with either dorado 0.5.3 or dorado 0.7.0 with parallel mapping to relevant reference (with option –secondary=no to exclude secondary alignments) and poly(A) detection turned on with the option –estimate-poly-a. Basecalled bam files were demultiplexed with dorado demux and sorted with samtools sort. Poly(A) lengths for each sequencing read were extracted from the pt:i tag (in the basecalled bam file) with a Python script. In the case of mRNA-1273 samples, the read sequence was also used for identifying the terminal pentamer from the basecalled sequence using regular expressions.
Statistical analysis of poly(A) lengths
P values for each transcript were estimated using the Kruskal–Wallis test and adjusted for multiple comparisons using the Benjamini–Hochberg method. Transcripts were considered as having a significant change in poly(A) tail length if the adjusted P value was less than 0.05 and if there were at least 20 supporting reads for each condition.
Differential expression analyses
Illumina RNA-seq reads were mapped to the mouse reference genome (GRCm38, ENSEMBL, release 94) using the STAR aligner (v.2.7.6a)39. Read counts were assigned to genes using featureCounts from the Subread package (v.2.0.1) with options -Q 10 -p -B -C -s 2 -g gene_id -t exon and the respective annotation file (Gencode v.M25). Multimappers and reads overlapping multiple features were not counted. Nanopore read counts were derived from the nanopolish polya output files, used for the analyses of poly(A) tail lengths. Reads assigned for each gene were summarized and obtained counts were used for subsequent analyses.
Differential expression analysis was performed with the DESeq2 (v.1.22) Bioconductor package40, using a likelihood ratio test for data from time-course experiments. Pair-wise comparisons of each time point were calculated with the Wald test, with log fold change values shrunk using the apeglm method. Genes with similar expression patterns were clustered with hierarchical clustering using the ward.D method and z-scored expression values for each gene. Heat maps were drawn with the ComplexHeatmap package41. g:Profiler42 or ClusterProfiler were used for Gene Ontology (GO) enrichment analysis. Genes were assigned as immune response genes (Fig. 4c and Supplementary Tables 6 and 7) if they were annotated to GO (Biological Process) terms GO:0140374, GO:0090714, GO:0034340, GO:0034341, GO:0034342, GO:0045087 or GO:0035455.
Codon usage analysis
Codon usage indices were analysed using the coRdon (https://github.com/BioinfoHR/coRdon) and cubar (https://github.com/mt1022/cubar) R packages. To compute the codon adaptation index, a reference set of 500 highly expressed genes in mBMDMs, based on a previous study7, was provided. For calculating the effective number of codons, codon bias or preference within the vertebrate context was used. The frequency of optimal codons was determined on the basis of the occurrence frequency of each amino acid in reference sequences. The mRNA vaccines BNT162b2 and mRNA-1273 were compared against the SARS-CoV-2 spike protein reference sequence from GenBank (accession: NC_045512.2:21563-25384), mouse endogenous transcripts (protein-coding), transcript groups selected by GO terms (for example, associated with the membrane or endoplasmic reticulum) and potential substrates of TENT5A and TENT5C, selected as per a previous study7, as well as IVT reporter sequences for OVA, PfCSP and ZikVE. GC content and other sequence features were assessed using functions available from Bioconductor and the GenRCA Rare Codon Analysis Tool (https://www.genscript.com/tools/rare-codon-analysis), providing mouse as the host organism and the Kazusa database as the codon usage reference. The results of the analyses were visualized with ggplot2.
Statistics and reproducibility
No statistical methods were used to predetermine sample sizes. Statistical analysis was performed on data from two or more biologically independent experimental replicates using the R environment or Prism 6 software unless otherwise stated. The statistical tests used in each instance are provided in the figure legends.
Data from the analyses of poly(A) tail length are presented as scatter dot plots, as indicated in the figure legends, and individual data points are shown. The median of poly(A) length is shown on the plots (as a horizontal bar and a numeric value) for mRNA populations with processed poly(A) tails (that is, without the terminal pentamer in the case of mRNA-1273, OVA, PfCSP and ZikVE reporters, or for all the reads in the case of BNT162b2); the mean value would be biased because poly(A) lengths do not follow a normal distribution. Plots indicate fractions of reads with elongated (blue-shaded) or shortened (red-shaded) tails. Classification thresholds (deadenylation and re-adenylation) were estimated as values close to the 0.2 and 0.8 quantiles (respectively) from crude vaccine RNA (with pentamer in the case of mRNA-1273). In the case of DRS data, the thresholds were set at 85 and 115 for mRNA-1273 and 95 and 125 for BNT162b2. For cDNA sequencing analysis with dorado, which usually gives shorter estimates of poly(A) tail length, the thresholds were set at 80 and 110 for mRNA-1273 tails and 50 and 80 for BNT162b2 tails (analysis was performed without additional segmentation, so the length of the last poly(A) segment was reported).
Most of the experiments were repeated at least twice, leading to comparable results, with the exception of the mRNA-1273 treatment of A549 cells and the viability assays, which were repeated once. Samples with clear technical failures during tissue collection, cell isolation, processing or data collection were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.