Mouse lines and treatments
Tie2-cre (Tg(Tek-cre)12Flv)53, Vegfr3-creERT2 (Flt4tm2.1(cre/ERT2)Sgo)21 Prox1-creERT2 (Tg(Prox1-cre/ERT2)1Tmak)28, R26–iMb2-Mosaic (Gt(ROSA)26Sortm1(CAG-EYFP*,-mEYFP*,-tdTomato*,-mTFP1*)Ben)20, Cldn5flox (Cldn5tm1c(EUCOMM)Wtsi)23, Cdh5-GFP (encoding VE-cadherin-GFP fusion protein)16, Cdc42flox (Cdc42tm1Brak)54 and Itgb1flox (Itgb1tm1Efu, The Jackson Laboratory, stock number 004605)55 mice were previously described, and analysed on a C57BL/6J background, with the exception of the R26-iMb2-Mosaic;Vegfr3-creERT2 mice used for intravital imaging experiments that were crossed to a C57BL/6-albino (B6(Cg)-Tyrc-2J/J) background. R26-LifeAct-EGFP (Gt(ROSA)26Sortm1(CAG-EGFP)Tmak) mice were generated as described in the Supplementary Information. Cre-mediated recombination in R26-LifeAct-EGFP mice was induced by topical application of 50 μg of 4-hydroxytamoxifen (4-OHT, H7904, Sigma-Aldrich) dissolved in acetone (10 mg ml−1) to the dorsal side of each ear. Cre-mediated recombination in R26-iMb2 mice, and gene deletion in mice carrying floxed alleles were induced by three (Cldn5 or Cdc42) or five (Itgb1) consecutive administrations of 1 mg of tamoxifen (T5648, Sigma-Aldrich) dissolved in peanut oil (10 mg ml−1, P2144, Sigma-Aldrich), by oral gavage. Littermate controls were included in each experiment, which were tamoxifen-treated Cre− mice or Cre+ mice carrying a heterozygous floxed allele. Increased interstitial fluid volume was induced by intradermal injection of 20 µl of sterile PBS into the ear of sedated mice, and ears were processed after 10 min for further analysis. Experimental procedures on mice were approved by the Uppsala Animal Experiment Ethics Board (permit numbers 130/15, 5.8.18-06383/2020 and 5.8.18-0336/2021) or the National Animal Experiment Board in Finland (licence number ESAVI/15852/2022) and performed in compliance with all relevant national regulations.
Antibodies
The details of primary antibodies used for immunofluorescence of whole-mount tissues and cells are provided in Supplementary Table 1. Secondary antibodies conjugated to Dylight405, AF405, AF488, AF555, AF594, AF647, AF680, Cy3 or horseradish peroxidase were obtained from Jackson ImmunoResearch Secondary antibodies conjugated to AF405+, AF488+, AF555+, AF594+ or AF647+, were obtained from Thermo Fisher Scientific. Actin was visualized using Phalloidin conjugated to AF647 (A22287, Invitrogen) or SPY-555 actin (SC202, Spirochrome). All were used in 1:200–1:1,000 dilution. Nuclei were visualized using 4,6-diamidino-2-phenylindole (DAPI) (1:1,000 in PBS). Fluorescent protein expression in the iMb2-Mosaic line was visualized using antibodies against HA epitope tag, dsRed or GFP.
Culture and isotropic stretch of primary ECs
Primary human dermal LECs (HDLECs) from juvenile foreskin (C-12216, PromoCell) and primary HUVECs (C-12200, PromoCell) were maintained on fibronectin-coated (2 μg ml−1; F1141-2MG, Sigma) cell culture dishes at 37 °C and 5% CO2. The cell lines were authenticated based on morphology and immunohistochemistry profile and tested negative for mycoplasma contamination. HDLECs were supplied with complete Endothelial Cell Growth Medium 2 (ECGMV2; C-22022, PromoCell) and HUVECs with complete ECGMV (C-22010, PromoCell). Cells were passaged up to six times using Trypsin-EDTA (25300054, Thermo Fisher Scientific), diluted to 0.025% with Dulbecco’s PBS (DPBS), before being used for the stretching experiments. The autoclaved PDMS chambers were functionalized for cell culture usage with 0.5 mg ml−1 Sulpho-SANPAH, diluted in sterile MilliQ water, under ultraviolet light for 10 min. The functionalized chambers were extensively washed in DPBS (14190-094, Gibco) and coated with fibronectin before seeding of cells (3 × 104 cells per mm2). 72 h after cell seeding, the chambers were transferred into 2% prestretch holders and cells were supplied with the complete ECGMV or ECGMV2 supplemented with 0.1% v/v Pen Strep (15140-122, Gibco) and 1 mM HEPES Buffer Solution (15630-056, Gibco). After 12 h, the chambers were transferred into the MultiStretcher device, developed as an extension of the IsoStretcher system22 to enable a simultaneous in-plane isotropic stretch of four custom-moulded PDMS chambers in parallel as described in the Supplementary Information. Details of the MultiStretcher device will be presented elsewhere (F.L., S.S. and O.F., in preparation). Cyclic isotropic stretching corresponding to roughly 12% change in membrane area was applied at 0.01 Hz (100 s per stretch cycle) for 14 h or 22 h at 37 °C and 5% CO2. Unstretched chambers, kept in prestretch holders, were used as a control. Young’s modulus of the PDMS substrate was measured as 2,391 ± 95 kPa (ref. 56).
For integrin β1 inhibition, HDLECs were treated at the onset of stretching with 0.1 or 0.2 μg ml−1 rat anti-human CD29 (mAb13) (552828, BD Pharmingen). For pharmacological inhibition of CDC42, HDLECs were treated at the onset of stretching with 15 μM ML141 (SML0407-5mg, Sigma) or vehicle (DMSO, 276855-100ML, Sigma-Aldrich). For silencing of CDC42 expression, HDLECs were first grown to confluence in fibronectin-coated PDMS chambers. Then 34 h before the start of the 14 h long stretching experiment, cells were transfected with either negative control (462001, Invitrogen) or CDC42 Stealth (HSS190761, Thermo Scientific) siRNA at the final concentration of 20 nM using Lipofectamine RNAiMAX Transfection Reagent (56531, Invitrogen) according to the manufacturer’s instructions.
Immunofluorescence
HDLECs and HUVECs were fixed inside the stretch chambers (in stretched state) with ice-cold 4% paraformaldehyde (PFA) in PBS for 10 min, followed by washing in PBS and permeabilization and blocking in 0.2% IGEPAL CA-630 (18896-50ML, Sigma) plus 3% bovine serum albumin (A3295, Sigma-Aldrich) in PBS for 1 h at room temperature. Primary antibodies dissolved in cell blocking buffer were added on cells and incubated overnight at 4 °C. After washing in PBS, cells were incubated with secondary antibodies and SPY-555 actin for 3 h at room temperature. Nuclear staining was done using DAPI (MBD0015-1ML, Sigma-Aldrich) in PBS for 10 min at room temperature, followed by further washing and imaging. To maintain the active conformation of integrin β1, HDLECs stained for active integrin β1 were washed with 1 mM MgCl2 in ice-cold DPBS and then fixed inside the chambers with ice-cold 4% PFA plus 1 mM MgCl2 in PBS for 10 min.
Optogenetic activation of CDC42
Lentiviral particles for Lck-mTurquoise2-iLID and SspB-HaloTag-ITSN1(DHPH) were produced in human embryonic kidney 293T cells (CRL-3216, American Tissue Culture Collection) as described in ref. 34. The cell line was authenticated based on morphology and tested negative for mycoplasma contamination. The HDLECs were transduced with both lentiviruses 6 days before imaging, and puromycin was added 3 days before imaging. Transduced HDLECs were subsequently seeded onto glass-bottom 12-well culture plates (ø14 mm, MatTek Corporation), coated with fibronectin and grown into confluent monolayers in complete ECGM2-MV medium. During imaging, HDLECs were grown in microscopy medium (20 mM HEPES (pH 7.4), 137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2 and 20 mM glucose) at 37 °C and 5% CO2. The HaloTag was stained 3 h before imaging, with a concentration of 150 nM of Janelia Fluor Dye (JF) JF552nm (red) (Janelia Materials). The medium was replaced before imaging. VE-cadherin was stained by adding Alexa Fluor 647 mouse anti-human CD144 (BD Pharmingen, 561567, 1:40) to the medium 2 min before the start of imaging. Details of the imaging parameters are provided in Supplementary Information. Before photoactivation, Lck-mTurquoise2-iLID was detected with a photomultiplier tube detector (gain 800 V), 447–523 nm emission detection range using a 442-nm diode laser line at 1% intensity. The 442-nm laser line was turned off during photoactivation experiments. Photoactivation was achieved by scanning the defined region of interest with a 488-nm laser line, the argon laser power set to 15% and intensity 5%, every 2.58 s for a total of 2.5 min. During photoactivation, SspB-HaloTag-ITSN1(DHPH) stained with JF552nm was detected with a photomultiplier tube detector (gain 750 V), 566–629 nm emission detection range with a DPSS 561-nm laser line at 1% intensity. Sequentially, VE-cadherin-Alexa Fluor 647 signal was detected using a HyD detector (gain 50%), 652–775 nm emission detection range in combination with a 647-nm HeNe laser line at 3% intensity.
Resistance measurements were performed using electrical cell-substrate impedance sensing (ECIS) ZTheta (Applied BioPhysics) at 4,000 Hz, representing paracellular permeability, every 10 s at 37 °C and 5% CO2. ECIS arrays (eight-well, ten-electrode, 8W10E PET) were pretreated with 10 mM cysteine (Sigma) for 15 min at 37 °C, washed twice with 0.9% NaCl solution and coated with 10 µg ml−1 fibronectin in 0.9% NaCl solution (Sigma) for at least 1 h at 37 °C. Lentivirally transduced HDLECs were seeded at 50,000 cells per well density to grow into a monolayer, and measurements were started immediately after seeding (n = 4 wells per condition). For global photoactivation, started roughly 22 h after seeding, an RGB LED safety strip (Combo 12 V/24 V SMD 3528/50505, Fuegobird) was taped to the lid of the cell culture dish with a rough distance of 1 cm from the cell monolayer and set to blue light (peak 470 nm, highest intensity setting 9) for 45 min. The corresponding blue LED light spectrum was measured as described in ref. 34.
Whole-mount immunofluorescence
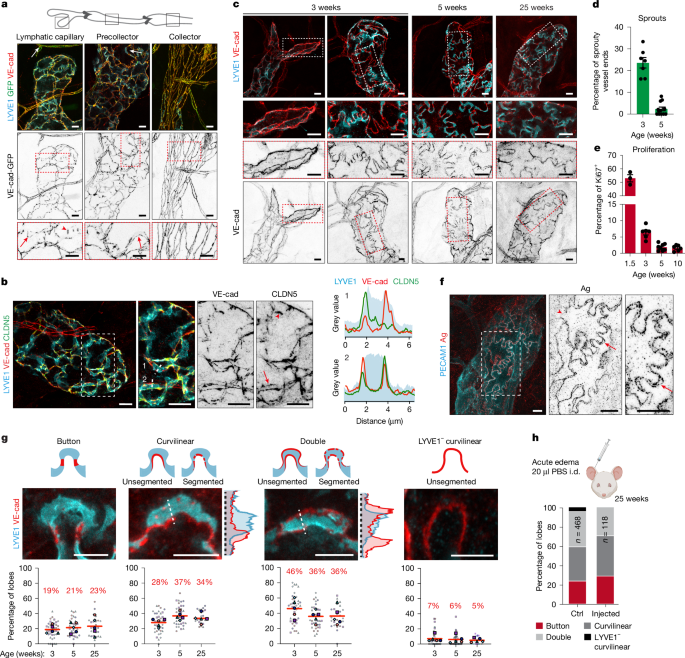
Most tissues (juvenile and adult ear skin, adult diaphragm or embryonic back skin) were harvested from mice that were euthanized by cervical dislocation or CO2 asphyxiation, and immediately dissected and placed for fixation in 4% PFA for 2 h at room temperature or 4 h at 4 °C. Tissue from Cdh5-GFP (Fig. 1a,b and Extended Data Fig. 1a) and Itgb1flox (Extended Data Fig. 6d,e) mice was harvested after transcardial perfusion following three different protocols, yielding a similar diversity of LEC junctions compared to immersion fixation (Supplementary Fig. 2c): (1) perfusion with 10 ml of PBS, followed by 10 ml of 4% PFA (room temperature), ear collection and postfixation by immersion in 4% PFA overnight at 4 °C, (2) perfusion with Hanks balanced salt solution, followed by 4% PFA, ear collection and postfixation by immersion in 4% PFA for 4 h at 4 °C and (3) direct perfusion with 1% formaldehyde for 2–3 min, ear collection and postfixation by immersion in 2% PFA for 4 h at 4 °C. Skin on the dorsal side of the ear pinna was dissected from the underlying cartilage layer before (immersion) or after (perfusion) fixation. Tissues were permeabilized in 0.3% Triton X-100 in PBS (PBST) for 10 min. After blocking in PBST with 2% bovine serum albumin and 1% FBS for 2 h, tissues were incubated with primary antibodies in blocking buffer overnight, followed by PBST washing and incubation with fluorescent dye-conjugated secondary antibodies for 2 h. All incubation steps were carried out at room temperature. Before mounting in Mowiol, samples were repeatedly washed in PBST and water. Details of consecutive staining of surface and total LYVE1, and staining using in vivo injected LYVE1 antibody and total LYVE1 as described in Supplementary Information.
Silver nitrate staining
Whole-mount AgNO3 staining was performed on immersion-fixed ear skin. Tissue was first blocked in blocking buffer (3% bovine serum albumin, 1% fetal bovine serum in TBS (Tris-buffered saline)) and subsequently stained using primary antibodies overnight at 4 °C dissolved in blocking buffer. The next day, tissue was washed in TBS and incubated with secondary antibodies dissolved in blocking buffer for 2 h at room temperature. Tissue was washed again in TBS for 2 h and was further processed for silver nitrate staining. In brief, tissue was washed twice for 1 min in d-glucose solution (280 mM), followed by immersion for 1 min in freshly filtered AgNO3 solution (15 mM) protected from light. After washing for a further 2 min in fresh glucose solution, the tissue was mounted and immediately underwent imaging. Silver stain was slowly developed under the microscope by shining white light using the microscope’s brightfield function until the desired contrast was reached. Sequential channel imaging was performed, whereby silver particles were imaged using far-red (685 nm) reflected light followed by antibody fluorescent signals (AF594 and AF555, respectively). See Supplementary Information for details about the specificity of silver staining.
Confocal microscopy and image processing
Confocal images were obtained using a Leica SP8 or Leica Stellaris 5 confocal microscope. Details of lasers and objectives are described in Supplementary Information. Images were deconvolved with Huygens Essential software (v.19.04) (Scientific Volume Imaging) (Figs. 3b,e, 4b and 5f–h, Extended Data Figs. 9d, 10a,b and 11a) or Leica Lightning (Figs. 2h and 4g,i, Extended Data Figs. 2b, 4a and 6b and Supplementary Fig. 6e), which is part of LasX software. Huygens deconvolution was used by using a theoretical point spread function and automatic background estimation. Stopping criteria were set to 40 iterations and a signal-to-noise ratio of 10. Lighting deconvolution was used using an adaptive approach, using the fitting optical parameters in terms of objective lens, corresponding wavelength. Maximum iterations were set to 20 with smoothing. Further processing was done using Fiji and ImageJ. Images represent maximum intensity projections of individual Z-stacks unless indicated otherwise. Processing of time-lapse intravital imaging videos was performed in Fiji and ImageJ. Individual frames were stabilized and 3D drift corrected using the image registration plug-in. To remove noise, a rolling average algorithm was used, part of the Multi Kymograph plug-in, which averages three subsequent frames.
Intravital multiphoton microscopy
Animals undergoing intravital imaging were sedated with an intraperitoneal injection of 100 mg kg−1 ketamine and 12.5 mg kg−1 xylazine dissolved in sterile saline. The dorsal ear skin was immobilized on a custom-made 3D printed stage for imaging. Animals received eye cream and thermal support during the entire imaging session, and those undergoing longitudinal imaging, were rehydrated using an intraperitoneal injection of saline after imaging. Imaging was performed using a LEICA SP8 DIVE platform equipped with a Ti:Sapphire multiphoton laser emitting a 680–1,300 nm tunable and 1,045 nm fixed laser line. All imaging was done using a HC IRAPO ×25/1.0 numerical aperture (NA) objective.
Image quantification
Details of image quantification are provided in Supplementary Information. Images of annotated lymphatic capillary junctions in mouse ear skin used for Fig. 1g are available at Zenodo (https://doi.org/10.5281/zenodo.13880404)57. Four categories were defined: (1) button junction, a punctate VE-cadherin+ deposit at the neck of LYVE1+ lobe/overlap, with no detectable VE-cadherin at the borders of the overlap; (2) curvilinear junction, unsegmented or segmented distribution of VE-cadherin within one border of LYVE1+ lobe/cellular overlap; (3) double junction, unsegmented or segmented distribution of VE-cadherin within both borders of LYVE1+ lobe–cellular overlap; (4) LYVE1− curvilinear junction, unsegmented linear VE-cadherin distribution at cell–cell contacts in the absence of LYVE1 and (5) zipper junction, continuous linear VE-cadherin distribution surrounding the entire cell in the absence of LYVE1.
Dextran clearance assay
Animals undergoing dextran clearance assay were sedated with an intraperitoneal injection of 100 mg kg−1 ketamine and 12.5 mg kg−1 xylazine dissolved in sterile saline. Animals received eye cream and thermal support during the entire imaging session. The dorsal ear skin was immobilized on a custom-made 3D printed stage for imaging and 1 µl of tracer solution containing 5 mg ml−1 TRITC-conjugated dextran (150 kDa, FD150, Sigma-Aldrich) was injected intradermally using a 32 G needle size Hamilton syringe. The entire ear was imaged using a HCX PL FLUOTAR ×5/0.15 NA objective within 3 min and animals were allowed to recover. After 4 h, animals were resedated and underwent a second round of imaging using the same imaging parameters to determine the clearance of injected tracer.
TEM
Mice were euthanized by cervical dislocation or CO2 asphyxiation. Skin on the dorsal side of the ear pinna was immediately dissected from the underlying cartilage layer, transferred to 2.5% glutaraldehyde (Ted Pella) + 1% PFA (Merck) in 0.1 M phosphate buffer pH 7.4 and incubated in the fixative at 4 °C overnight. Fixed ears were cut longitudinally along the proximal–distal axis into 2–3 mm strips and further incubated in fresh fixative, which was replenished once more before dehydration. The strips from the central region of the ear were further divided into three parts in the proximal–distal axis. Only the distal part close to the tips of the ear that lacks larger collecting vessels and contains a plexus of lymphatic capillaries and precollecting vessels composed of oak leaf-shaped LECs was used for analysis. Before sectioning, samples were washed in 0.1 M phosphate buffer, incubated in 1% osmium tetroxide in 0.1 M phosphate buffer for 1 h, rinsed again, dehydrated in increasing concentrations of ethanol (50, 70, 95 and 99.9%) for 10 min, and incubated in propylene oxide for 5 min. Samples were then placed in Epon Resin (Ted Pella) and propylene oxide (1:1) for 1 h, followed by 100% resin overnight. Subsequently, samples were embedded in capsules in newly prepared Epon Resin and after 1–2 h finally polymerized at 60 °C for 48 h. Sectioning was carried out using an EM UC7 Ultramicrotome (Leica). Then 60–70-nm-thin sections were placed on a grid and contrasted with 5% uranyl acetate and Reynold’s lead citrate. Imaging was carried out on a Tecnai G2 Spirit BioTwin transmission electron microscope (Thermo Fisher/FEI) at 80 kV with an ORIUS SC200 CCD camera and Gatan Digital Micrograph software (both Gatan Inc.).
scRNA-seq data analysis
Differentially expressed genes between the different LEC subtypes were visualized using a Shiny-based web application for dermal mouse LECs25 available at https://makinenlab.shinyapps.io/DermaLymphaticEndothelialCells/.
FEM simulations
The FEM simulations were performed with MorphoMechanX using available models adapted from ref. 27. A regular cylindrical grid of 45 µm wide and 200 µm long was created and outlines from the cells of a lymphatic vessel obtained from confocal imaging were projected onto it and smoothed. These cells were then extruded inward to generate 3D volumetric cells with a depth of 2 µm and triangulated using a threshold area of 4 µm. The template was used as the reference configuration for triangular three-node membrane elements that were given a thickness of 0.1 µm. An isotropic St. Venant material model (linear, large deformation) was used with the Young’s modulus set to 100 kPa to match a 10-kPa cell level Young’s modulus estimated from the literature for ECs30,31,32 (ignoring the cell ends, the 2 × 0.1 µm membrane thickness occupied roughly one-tenth the cross-sectional area of the cells that were 2 µm deep). A uniform internal pressure was applied normal to the inside faces of the elements, which cancels out on the shared walls between cells. For simulations with a lower pressure inside the vessel, the inside faces were assigned a higher pressure. Stresses were visualized as the trace of the stress tensor.
Statistics
Graphpad Prism (v.9) was used for graphic representation of the data. No statistical methods were used to predetermine sample size. For in vivo experiments, a minimum of three mice per condition was used, except for Figs. 2j and 4c,d, n = 2 for 9-week-old mice. For in vitro experiments a minimum of three biological replicates were used, except for Extended Data Fig. 10a (validation of integrin beta1 inhibition), n = 1–2 stretch holders. The sample size of three was chosen as the minimum required to perform statistical tests. Allocation of mice into experimental groups was based on genotype. Data were collected from different litters on different days and experiments were performed for different batches at different time points. For in vitro experiments, allocation into experimental groups was performed randomly. No blinding was done in the analysis and quantifications. Data between two groups were compared using an unpaired two-tailed Student’s t-test assuming equal variance. When the data were not normally distributed Mann–Whitney U-test was used instead. Data between multiple groups were compared using an ordinary one-way analysis of variance (ANOVA) or Brown–Forsythe and Welch ANOVA followed with multiple testing, and categorical variables were compared using Fisher’s exact test. Differences were considered statistically significant when P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.