Human specimens and clinical data
PDAC tissues were obtained from the General and Pancreatic Surgery Unit at the University of Verona. Written informed consent was obtained from patients preceding the acquisition of the specimens. The fresh tissues used to establish PDOs and associated clinical and follow-up data were collected under a study approved by the Integrated University Hospital Trust (AOUI) Ethics Committee (Comitato Etico Azienda Ospedaliera Universitaria Integrata): approval number 1911 (protocol number 61413, Prog 1911 on 19 September 2018). FFPE tissues were collected under protocol number 1885 approved by the AOUI Ethics Committee and retrieved from the ARC-NET Biobank.
PDO establishment and culture
PDAC PDOs were established following previously published procedures17. The specimens used to generate PDOs were examined by pathologists to confirm the presence of neoplastic cells. In brief, tissue specimens were minced and digested with collagenase II (5 mg ml−1; Gibco) and dispase I (1.25 mg ml−1; Gibco) in human splitting medium (HSM; advanced Dulbecco’s modified eagles medium with nutrient mixture F-12 Hams (Gibco) supplemented with HEPES (10 mM; Gibco), Glutamax (2 mM; Gibco) and Primocin (1 mg ml−1; InvivoGen)) at 37 °C for a maximum of 2 h, followed by an additional 15-min digestion with TrypLE (Gibco) at 37 °C. The digested material was embedded in growth factor-reduced Matrigel (Corning) and overlaid with human complete medium (+WR; mouse epidermal growth factor (50 ng ml−1; Gibco), B-27 Supplement (1X; Gibco), nicotinamide (10 mM; Sigma-Aldrich), N-acetylcysteine (1.25 mM; Sigma-Aldrich), FGF10 (100 ng ml−1; Peprotech), Y-27632 dihydrochloride (10.5 µM; Sigma), gastrin (10 nM; Tocris), TGFβ receptor inhibitor A83-01 (500 nM; Tocris), WNT3A-conditioned media (50% v/v), RSPO1-conditioned media (10% v/v) and mouse Noggin (100 ng ml−1; Peprotech)). Media were refreshed every 3–4 days. For organoid propagation, confluent organoids were removed from Matrigel, dissociated into small clusters of cells by pipetting, and resuspended in an appropriate volume of fresh Matrigel. All organoid models were acquired as part of the HCMI (https://ocg.cancer.gov/programs/HCMI) and are available for access from the American Type Culture Collection (ATCC). For each PDO, Supplementary Table 1 provides two unique identifiers (study ID and HCMI ID), along with the clinical and follow-up data associated with the corresponding case. The HCMI ID can be queried in the HCMI searchable catalogue (https://hcmi-searchable-catalog.nci.nih.gov/). Dependency of organoid cultures to WNT3A and RSPO1 was assessed on nine PDOs (VR01, VR02, VR06, VR09, VR20, VR21, VR23, VR29 and VR32). Organoid cultures were passaged once a week with a splitting ratio of 1:3 in +WR or human-depleted media (−WR; mouse epidermal growth factor (50 ng ml−1; Gibco), B-27 supplement (1X; Gibco), nicotinamide (10 mM; Sigma-Aldrich), N-acetylcysteine (1.25 mM; Sigma-Aldrich), FGF10 (100 ng ml−1; Peprotech), and gastrin (10 nM; Tocris)). To establish WRi PDOs, organoids established and propagated in +WR were placed and maintained in −WR until the emergence of WRi. Owing to the cell death induced by −WR, the media were refreshed every 3 days and Matrigel every 14 days without propagating the cultures, until the emergence of WRi PDOs. The growth curve of WRi PDOs was obtained by plotting the number of domes (one dome refers to 50 μl of Matrigel) at different days of culture in −WR. WRi PDOs were reintroduced in +WR or maintained in –WR (control) for five passages before collection of metaphase spreads and proteins. To obtain ‘late-passage’ PDOs, organoids were passaged 40 times post-establishment in +WR medium. For the Wnt-C59 experiment, baseline and adapted organoids were passaged every 7 days with a splitting ratio of 1:3 in the presence of Wnt-C59 (100 nM; Selleckchem). Wnt-C59 was added to the culture at the day of splitting and after 3 days of culture. Organoids were routinely tested for the presence of Mycoplasma contamination using the Mycoalert Mycoplasma Detection kit (Lonza).
Single-cell dissociation from organoids
Organoids were incubated with dispase I diluted in HSM (2 mg ml−1; dispase I solution) for 20 min at 37 °C to digest Matrigel. Following this, organoids were dissociated using TrypLE (Gibco) for 10 min at 37 °C, incubated in dispase I solution for additional 10 min at 37 °C, and pipetted to obtain single-cell suspension.
Assessing MYC activation by WR media
PDOs were dissociated into single cells as previously described and plated in Matrigel in +WR (100,000 viable cells per condition). Following organoid reformation in +WR, PDOs were starved overnight in HSM. Post-starvation, PDOs were stimulated with +WR, −WR or HSM for 8 h, before collection and isolation of RNA.
JQ1 in vitro treatment
Organoids were dissociated into single cells as previously described. One thousand viable cells were plated in 100 µl 10% Matrigel/media per well in a 96-well plate in triplicates. JQ1 (500 nM; S7110, Selleckchem) or vehicle was added 40 h after plating once the organoids were reformed. After 72 h of treatment, cell viability was assessed using CellTiter-Glo (Promega) following the manufacturer’s instructions. Results were normalized to the vehicle control of each PDO. In parallel, 20,000 viable cells per 50 μl Matrigel were plated and supplemented with media. Following organoid reformation, cells were treated with JQ1 (500 nM) or vehicle control, and RNA, and metaphase spreads were collected after 72 h.
Lentiviral production and infection of organoids
To overexpress MYC, we used a lentiviral vector carrying an open-reading frame for MYC (mGFP tagged; RC201611L4, Origene). Lentivirus was produced by transfecting the plasmid containing MYC, and the packaging plasmid VSV-G with X-tremeGENE9 (6365779001, Roche Sigma-Aldrich) in HEK293T cells. The viral supernatant was harvested 48 h post-transfection and quantified using the Lenti-XTM qRT–PCR Titration kit (Takara Bio) according to the manufacturer’s instructions. pLenti-C-MYC-DDK-P2A-Puro lentiORF control particles (PS100092V, Origene) were used as non-targeting control. The lentiviral barcoding library was produced by transfecting the plasmid library and the packaging plasmids pMD2.G and psPAX2 (gifts from D. Trono; Addgene plasmid #12259 and Addgene plasmid #12260) in HEK293T/17 cells (ATCC: CRL-11268). The viral supernatant was harvested 48 h post-transfection and concentrated with Lenti-X Concentrator (Clontech), according to the manufacturer’s instructions. Viral particles were resuspended in OPTI-MEM (Life Technologies), titrated using a fluorometric assay and stored at −80 °C. For infection, organoids were dissociated into single cells, resuspended in infection media (DMEM; Gibco), 5% fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin (Gibco), supplemented with 1 μg ml−1 polybrene and lentiviral particles. Cells were then spinoculated for 1 h at room temperature and incubated at 37 °C for 16 h. Infected cells were then collected, embedded in Matrigel and overlayed with +WR media. Antibiotic selection was started 48 h after infection using 2 µg ml−1 puromycin (Gibco).
Barcoding of organoids
For barcoding experiment, we used a 1M-barcode pool from the CloneTracker XP 3M Barcode-3′ Library in pScribe4-RFP-Puro (Cellecta) kit. We infected 5 × 105 cells with 0.1 multiplicity of infection of virus to obtain a population of cells with a single barcode per cell. After infection, organoids were subjected to antibiotic selection using 2 µg ml−1 of puromycin (Gibco). Barcoded organoids were then divided into two conditions: +WR (control) and −WR (selective pressure) with at least two replicates per condition. An aliquot of barcoded organoids was collected for DNA extraction (P0). The control condition was expanded for five passages before collection of the pellet, whereas pellets from the replicates in the presence of selective pressure were collected at the time of emergence of WR independence.
Organoid metaphase spreads and interphase nuclei
Organoids were incubated with Colcemid (1 µg ml−1; Gibco) in culture media at 37 °C and 5% CO2 overnight. Following incubation, organoids were dissociated into single cells as previously described. Single cells were incubated in hypotonic solution (potassium chloride 0.56% and sodium citrate 0.8%) for 20 min at room temperature. Nuclei were then fixed in ice-cold methanol–acetic acid (3:1), washed with methanol–acetic acid (2:1) and dropped on adhesion microscope slides.
DNA FISH
DNA FISH was performed using the ZytoLight SPEC MYC/CEN8 Dual Color FISH probe (ZytoVision). Before hybridization, tissues were deparaffinized and rehydrated, pre-treated with 0.1 citrate buffer (pH 6) solution at 85 °C for 30 min, followed by pepsin treatment (4 mg ml−1 in 0.9% NaCl, pH 1.5) for 4 min at 37 °C. For both tissues and PDOs, the probes were applied to the slides and sealed with rubber cement and incubated in a humidified atmosphere (Thermobrite System) at 80 °C for 10 min to allow denaturation of the probes and of the DNA target. Slides were then incubated overnight at 37 °C to allow for hybridization. The rubber cement and the coverslip were then removed, and the slides were washed in 2X SSC/0.3 % NP-40 for 15 min at room temperature and then at 72 °C for 2 min. Following post-hybridization washes, slides were counterstained with DAPI 1 μg ml−1 (Kreatech, Leica). For tissues and embedded organoids, images were acquired on a FV4000 confocal microscope (Olympus Life Science). Nuclei were acquired and visualized in blue (DAPI). For PDOs, images were acquired with Leica TCS SP5 Fluorescent microscopes. The number of fluorescent signals for each probe for each nucleus, for both tissues and PDOs, was quantified with FIJI (ImageJ2 v2.9.0/1.53t) using the Find plugin maxima function in a supervised manner as previously described6. Interphase ecDNA clustering was quantified by the autocorrelation function as described in Hung et al.28.
Histology and immunostaining
For histopathological analysis, organoids were released from Matrigel using dispase I solution as previously described, fixed with 10% neutral-buffered formalin for 20 min, and embedded in Histogel processing gel (Fisher Scientific). Histogel-embedded organoids were processed according to routine histology procedures and embedded in paraffin. To account for the effect of the media, +WR PDOs were put in −WR for 24 h before embedding and fixation. Haematoxylin and eosin (H&E) staining and immunostainings were performed on sections of FFPE tissues and organoids, following established procedures using the reported primary antibodies: MYC (clone EP121, ALI415G7, Biocare for immunohistochemistry (IHC); clone Y69, ab32072, Abcam for immunofluorescence, dilution 1:500), GATA6 (polyclonal; AF1700, R&D Systems for IHC, dilution 1:200), ΔNp63 (clone BC28, PA0163, Leica for IHC), CK5 (clone XM26, PA0468, Novocastra for IHC, dilution 1:100) and γH2AX (clone CR55T33, 14-9865-82, eBioscience for immunofluorescence, dilution 1:500). For immunofluorescence, we used the following secondary antibody: Alexa Fluor 488 donkey anti-mouse (A21202, lot 1423052, Invitrogen, dilution 1:500), Alexa Fluor Plus555 goat anti-rabbit (A32732, lot VC297826, Invitrogen, dilution 1:500). Immunohistochemistry slides were then scanned and digitalized using the Aperio Scan-Scope XT Slide Scanner (Aperio Technologies). In tissues, quantification of MYC staining was performed on 20 areas for each tissue and reported as the H-score. In organoids, MYC and GATA6 staining was quantified as the percentage of positive nuclei per organoid, using Aperio ImageScope. For immunofluorescence, images were acquired by the FV4000 confocal microscope (Olympus Life Science) and quantified using ImageJ (https://imagej.nih.gov/).
ImmunoFISH
Single-cell suspension from baseline PDOs was obtained as previously described, then spun at 1,000 rpm for 5 min on slides using the Cytospin 4 cytocentrifuge and fixed by 4% paraformaldehyde (PFA) for 15 min. After washing in PBS, cells were permeabilized with 0.4% Triton for 10 min and incubated for 1 h in blocking solution (5% BSA, 5% goat serum and 0.1% Triton). After incubation with primary (MYC; ab32072, Abcam, dilution 1:500) and secondary (Alexa Fluor 647 goat anti-rabbit; A-21245, lot 2833435, Invitrogen, dilution 1:500) antibodies, cells were fixed again with 4% PFA for 10 min and washed with 2X SSC buffer. The ZytoLight SPEC MYC/CEN8 Dual Color FISH probe (ZytoVision) was applied as previously described. Images were acquired with a FV4000 confocal microscope (Olympus Life Science) and quantified using ImageJ (https://imagej.nih.gov/).
Pulse EdU staining
Baseline and WRi organoids were cultured for 3 days in +WR and −WR. At day 3, fresh media with EdU (10 μM) was added for 1 h before organoid collection and dissociation into single-cell suspension. Cells were spun on slides, fixed with 4% PFA and washed with 3% BSA in PBS. EdU detection was performed following the manufacturer’s instructions (Click-iT EdU imaging kit Alexa Fluor 555, C10338, Invitrogen), followed by a second fixation with 4% PFA for 10 min. FISH was performed as previously described using the ZytoLight SPEC MYC/CEN8 Dual Color FISH probe (ZytoVision). Images were acquired with a FV4000 confocal microscope (Olympus Life Science) and quantified using ImageJ (https://imagej.nih.gov/).
RNAScope
MYC mRNA in situ detection on embedded organoids was performed using the RNAScope Multiplex Fluorescent Reagent kit v2 (323100, Bio-Techne) following the manufacturer’s instructions. The target probe Hs-MYC-C2 (311761-C2, Bio-Techne) together with the Opal 570 fluorophores were used. The nuclei were counterstained and visualized using DAPI fluorescent dye. Images were acquired with a FV4000 confocal microscope (Olympus Life Science) and quantified using ImageJ (https://imagej.nih.gov/).
Immunoblotting
Proteins were prepared using cell lysis buffer (Cell Signaling Technology) supplemented with protease inhibitor cocktail (Sigma) and the phosphatase inhibitor PhosphoSTOP (Roche). Protein lysates were separated on 4–12% Bis-Tris NuPAGE gels (Life technologies), transferred to a PDVF membrane (Millipore) and then incubated with the reported antibodies: MYC (clone Y69, ab32072, lot 1012026-1, Abcam, dilution 1:1,000), γH2AX (clone EP854(2)Y, ab81299, lot GR3203642-4, Abcam, dilution 1:1,000), GFP (clone D5.1, 2956, lot 6, Cell Signaling Technologies, dilution 1:1,000), GAPDH (clone D16H11, 5174, lot 6, Cell Signaling Technologies, dilution 1:5,000) and histone H3 (polyclonal, 09-838, lot 2698469, Sigma-Aldrich, dilution 1:5,000) primary antibodies, and the peroxidase-conjugated AffiniPure donkey anti-rabbit (polyclonal, 711-035-152, Jackson Immunoresearch Laboratories) secondary antibody. To account for the effect of the media, +WR PDOs were put in −WR for 24 h before collection of the pellet of the cells. For gel source data, see Supplementary Fig. 1.
Quantitative real-time PCR analysis
RNA from organoids were isolated using the TRIzol reagent (Life Technologies), followed by the column-based PureLink RNA Mini Kit (Thermo Fisher Scientific). Reverse transcription of 1 µg of RNA was performed using the TaqMan Reverse Transcription reagents (Applied Biosystems), and 20 ng of cDNA was used in the PCR. The following TaqMan probes HPRT1 (Hs02800695_m1) and LGR5 (Hs00173664_m1) were used. The following primers (Eurofins) were used with SYBR Green PCR master mix (Thermo Fisher): MYC forward: CCTGGTGCTCCATGAGGAGAG; MYC reverse: CAGACTCTGACCTTTTGCCAG; GAPDH forward: ACAGTTGCCATGTAGACC; GAPDH reverse: TTTTTGGTTGAGCACAGG.
Relative gene expression quantification was performed using the ΔΔCt method with the Sequence Detection Systems Software, v1.9.1 (Applied Biosystems).
DNA isolation
Organoids were incubated in Cell Recovery Solution (Corning) for 30 min at 4 °C to remove Matrigel, and were pelleted by centrifuging at 10,000g for 5 min at 4 °C. For tissues, slices from snap-frozen PDAC tissues were assessed by a pathologist for percent neoplastic cellularity, and only tissues with higher than 20% neoplastic cellularity were used. For WGS and panel DNA sequencing, DNA isolation was performed using the DNeasy Blood & Tissue kit (Qiagen). For CIRCLE-seq, high-molecular-weight DNA was extracted using the MagAttract HMW DNA Kit (Qiagen).
Whole-genome sequencing
DNA quality was assessed by DNF-467 genomic DNA 50 kb Kit on a Bioanalyzer 2100 (Agilent). Libraries were prepared and sequenced using NovaSeq 6000 S4 Reagent Kit v1.5 (300 cycles) at 15× coverage for 160 million reads per sample.
Data pre-processing and alignment
Sequencing data were pre-processed and mapped to the reference genome using the nf-core/sarek pipeline (v3.0.2)51. In brief, Fastp (v0.23.2)52 removed low-quality bases and adapters, BWA Mem (v0.7.17-r1188)53 mapped trimmed reads to the reference genome GRCh38 (v1.4.4), provided by the Genome Reference Consortium (https://www.ncbi.nlm.nih.gov/grc), mapped reads were marked for duplicates using Picard Markduplicates (v4.2.6.1), and read base-quality scores were recalibrated using GATK BaseRecalibrator (v4.2.6.1) and GATK ApplyBQSR (v4.2.6.1)54.
Amplicon characterization
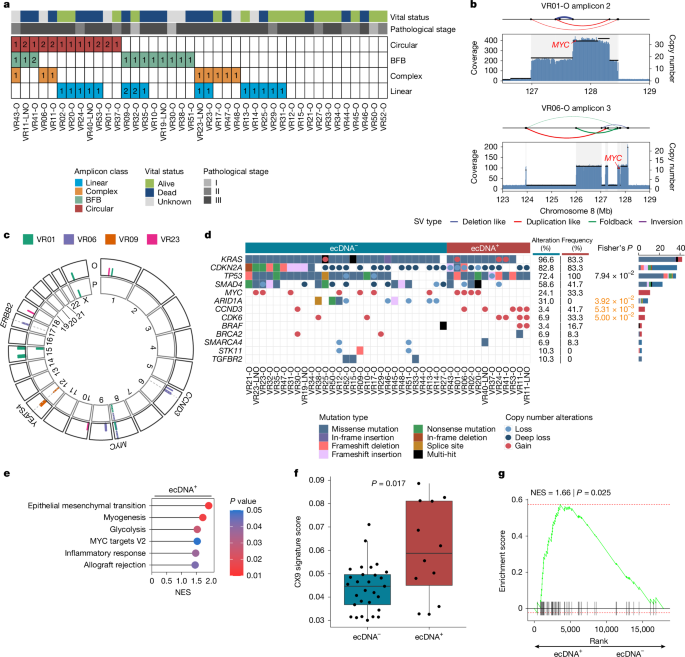
The nf-core/circdna (v1.0.1; https://github.com/nf-core/circdna) pipeline branch ‘AmpliconArchitect’ was used to define amplicon classes in each WGS sample. Nf-core/circdna calls copy number using cnvkit (v0.9.9)55 and prepares amplified segments with a copy number greater than 4.5 for AmpliconArchitect by utilizing the functionality of the AmpliconSuite-Pipeline (https://github.com/jluebeck/AmpliconSuite-pipeline). AmpliconArchitect (v1.3_r1)20 was ran on the aligned reads and the amplified seeds to delineate the amplicon structures. Identified amplicons were then classified using AmpliconClassifier (v0.4.11)56 into circular (ecDNA), linear (linear amplicon), complex (complex amplicon) or BFB (amplicon with a breakage-fusion-bridge signature). Samples containing at least one circular amplicon (ecDNA) were termed ‘ecDNA+’, whereas samples without ecDNA amplicons were termed ‘ecDNA−’. Samples were also classified into ‘circular’, ‘linear’, ‘complex’, ‘BFB’ or ‘no-fSCNA’ (no-focal somatic copy number amplification detected) by the types of amplicons they contained (see Kim et al.21). Samples with multiple amplicons were classified based on the amplicon with the highest priority. The priority is: circular > BFB > complex > linear. The amplicon similarity score and its P values were calculated based on the amplicon regions and breakpoint overlap as described in Luebeck et al.56.
Copy number calling
Copy number calls of the WGS samples were generated by cnvkit (v0.9.9)55. The identified segments were then classified as gain (copy number ≥ 3), loss (copy number ≤ 1) or deep loss (copy number ≤ 0.25).
Chromosomal instability signatures
Chromosomal instability signatures, including the CX9 replication stress signature, were assessed from the WGS copy number profiles using the R-package CINSignatureQuantification25.
Ploidy analysis
Sample ploidy was derived using PURPLE (v3.8.1)57, which estimates copy number and ploidy by using read depth ratio and tumour B allele frequency from COBALT (v3.9) and AMBER (v1.14), respectively (https://github.com/hartwigmedical/hmftools). COBALT, AMBER and PURPLE were used in tumour-only mode using their default parameters. Of note, PURPLE was used with a fixed parameter value of purity set to 1 for all samples, ensuring consistency in the analysis.
CIRCLE-seq
To enrich circular DNA for sequencing, each DNA sample was digested for 7 consecutive days with ATP-dependent Plasmid-Safe DNase (Lucigen) to remove linear/chromosomal DNA. Each day, 20 units of enzyme and 4 µl of a 25 mM ATP solution were added. After 7 days, the DNase was heat-inactivated for 30 min at 70 °C. The fold-change reduction in linear DNA was assessed by qPCR targeting the chromosomal gene HBB and the mitochondrial gene MT-CO1. Amplification of circular DNA was performed with a Phi29 polymerase as described in Koche et al.58. Amplified circular DNAs were then prepared for sequencing. In brief, around 550 ng of DNA was sheared to a mean length of around 400–450 bp and subjected to library preparation using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB), which included sequencing adapter addition and amplification. DNA Clean-up was performed using the Agencourt AMPure XP magnetic beads. All prepared libraries were sequenced using the Illumina NextSeq500 with the NextSeq 500/550 Mid Output Kit v2.5 (300 cycles), generating around 10 million paired-end 150-bp reads per sample.
Data processing
Sequencing reads were trimmed for both quality and adaptor sequences using cutadapt (v3.4)59. Trimmed reads were aligned to the GRCh38 reference genome using BWA Mem (v0.7.17-r1188)53.
Identification of sequencing coverage
Sequencing read coverage per 50-bp bin was calculated using deeptools ‘bamCoverage’ (v3.5.1)60 with default values. For visualization, the 50-bp read coverage values were combined into 10,000-bp bins using the function ‘ScoreMatrixBin’ of the genomation (v1.2.6) R package61.
Validation of ecDNA breakpoint
VR01 and VR06 breakpoint sequences were inferred by AmpliconArchitect and used to design the following primers: VR01 forward (5′>3′): TACATGGGGCTCTGCTACCTGC; VR01 reverse (5′>3′): AGCCTGTCCCTTTTCCCACCCA; VR06 forward (5′>3′): TGCCTGCTTGTGTGAACTTGGCT; VR06 reverse (5′>3′): AGGTGGTGGGGGAGGCCTAAAA.
Breakpoint regions were amplified by PCR containing 30 ng of gDNA, 2.5 μl of buffer (Quantabio), 0.5 μl of 10 mM NTP Mix (Thermo Fisher Scientific), 1 μl of primers and 0.25 μl of AccuStart II Taq DNA polymerase (Quantabio) in a total volume of 25 μl. The PCR amplification cycling conditions were 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s, with a final step at 72 °C for 5 min. PCR products were verified using the 5300 Fragment Analyzer (Agilent), purified using the ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific) and sequenced by capillary electrophoresis using the BigDye Terminator v3.1 (Thermo Fisher Scientific) on the Applied Biosystems 3500dx Genetic Analyzer (Thermo Fisher Scientific). Sequencing data were aligned to the human genomic DNA using Primer-BLAST.
Droplet digital PCR
ddPCR was conducted on a QX200 ddPCR system (Bio-Rad Laboratories). For VR01 and VR06, the probes targeting the copy number variations (CNVs) were designed on the interconnected breakpoint of each ecDNA using Bio-Rad Laboratories software (https://www.bio-rad.com/digital-assays/assays-create/cnd). The breakpoints were previously inferred through AmpliconArchitect and validated via Sanger sequencing (Extended Data Fig. 1h). The other probes used were commercially available (Bio-Rad Laboratories): MYC (FAM dHsaCP2500322), EPHA3 (FAM dHsaCP52506272) and TTC5 (HEX dHsaCP2506733) as reference genes for VR01 and VR06, and EPHA3 (FAM dHsaCP52506272) and PLEKHF1 (HEX dHsaCP2506723) as reference genes for VR23. Amplification was performed using the ddPCR Supermix for Probes, following the manufacturer’s instructions (Bio-Rad Laboratories). Each reaction used 1 ng of genomic DNA in 20 μl of volume, containing probes (from 20X stock) and restriction enzyme (from 5 U μl−1). The reaction was partitioned into approximately 20,000 droplets by an automated droplet generator according to the manufacturer’s protocol (Bio-Rad Laboratories). The droplets were then transferred to a 96-well PCR plate and heat-sealed using the PX1 PCR plate sealer (Bio-Rad Laboratories). The PCR amplification cycling conditions were 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 60 °C for 60 s, and a final step at 98 °C for 10 min. After thermal cycling, each droplet was scanned using the QX200 Droplet Digital PCR system (Bio-Rad Laboratories). Positive and negative droplets in each fluorescent channel (HEX and FAM) were distinguished based on fluorescence amplitude, using a global threshold set by the minimal intrinsic fluorescence signal resulting from the imperfect quenching of the fluorogenic probes in negative droplets, compared with the strong fluorescence signal from cleaved probes in droplets with an amplified template (or templates). The QuantaSoft (v1.3.2.0) software was used to analyse CNVs.
Barcode sequencing
Genomic DNA was isolated as previously described and quantified using Qubit 4 Fluorometer (Thermo Fisher Scientific). We amplified the barcodes from the CloneTracker XP library and added the Illumina adaptors as well as unique sample index sequences for multiplex sequencing using the NGS Prep Kit for Barcode libraries in pScribe (LNGS-300). The first PCR was performed using 25 ng of genomic DNA and the following cycling conditions: 30 cycles of 95 °C and 65 °C for 30 s each and 68 °C for 2 min. An aliquot of 5 μl from the first-round PCR was used for the second-round PCR that was performed under the following cycling conditions: 12 cycles of 95 °C and 65 °C for 30 s each and 68 °C for 2 min. After quantification by Fragment Analyzer High Sensitivity NGS kit (Agilent Technologies), we first combined PCR products from each sample at the same amount, then we purified and concentrated the pool using Qiagen QIAquick PCR Purification Kit following the manufacturer’s protocol. We quantified the library pools using NanoDrop spectrophotometer (Thermo Fisher Scientific) and then sequenced with custom read and index primers on the NextSeq2000 (Illumina), generating 150-bp paired-end reads at a depth of 5 million reads per sample. Of spiked-in PhiX control, 10% was sequenced by also adding the Standard Illumina primers to the custom read well.
Barcode sequencing analysis
Barcodes from genomic DNA sequencing were quantified using BWA (v0.7.18) and FeatureCounts from Rsubread package (v2.18.0). A FASTA reference file was first generated from the barcode pool provided by Cellecta. The sequencing reads were then aligned to the reference using the BWA-MEM (v0.7.17-r1188) algorithm. The resulting BAM files were subsequently used to quantify barcode abundances through the FeatureCounts functionality, as implemented in the R package Rsubread. To identify the presence of predominant barcodes, we selected those with a frequency exceeding 10% of the total barcode frequency within each condition.
DNA panel sequencing
Library preparation was performed using SureSelectXT HS Target Enrichment System (Agilent). Panel pair-end 2 × 150 sequencing was performed on NextSeq 550 (Illumina). Genes present in the panel are reported in Supplementary Table 3.
RNA sequencing
RNA from organoids were isolated using the TRIzol reagent (Life Technologies), followed by the column-based PureLink RNA Mini Kit (Thermo Fisher Scientific). Purified RNA quality was evaluated using the RNA 6000 Nano kit on a Bioanalyzer 2100 (Agilent), and only RNA with an RNA integrity number greater than 9 was used. The RNA sequencing library was obtained using poly(A) enrichment with the TrueSeq Stranded mRNA Library Prep kit (Illumina). Libraries obtained from PDOs at baseline (n = 14; analyses displayed in Fig. 1) were sequenced to a depth of 30 million fragments and 150-bp paired-end reads on an Illumina NextSeq 500 sequencer. For comparison between +WR and WRi PDOs, +WR PDOs were put in −WR for 24 h before RNA collection, to account for effect of the media. The resulting libraries were sequenced to a depth of 11 million fragments for organoids and 75-bp paired-end reads on an Illumina NextSeq 500 sequencer.
RNA sequencing analysis
Reads were aligned to the GRCh38 genome using STAR (v2.7), and the transcripts were quantified with RSEM (v1.3.3). For downstream analyses, the raw counts were normalized using the ‘rlog’ function of the DESeq2 R package. Genes with less than a total of 20 counts across all PDOs were removed before normalization. To compare gene expression values across amplicon types, the normalized gene values were Z-score normalized.
Tumour purity inference
The ESTIMATE (estimation of stromal and immune cells in malignant tumour tissues using expression data) tool was used to infer tumour purity of a subset of tumours from the ICGC (n = 50) and PDOs (n = 14) as previously described62.
Gene set enrichment analysis
Differential gene expression analysis was conducted using ‘DESeq2’ (v1.34.0)63. log2 Fold-change shrinkage was applied using the ‘lfcShrink’ function in DESeq2 with the ‘ashr’ method64. Gene set enrichment analysis was performed using the ‘fgsea’ R package (v1.20.0)65 with the Hallmark pathways database provided by the ‘msigdbr’ R package (v7.5.1)66.
Subtyping
Subtyping was performed scoring the samples according to the Raghavan signatures41 with the gene set variation analysis function (v1.42.0) and assigning the subtype according to what signature (basal or classical) achieved the highest score.
Fusion analysis
Fusion analysis was performed on adapted organoids to exclude the presence of chimeric proteins reactivating the WNT pathway. The nf-core/rnafusion (v3.0.0) pipeline was used to evaluate gene fusion from our RNA sequencing data; the pipeline was run under default parameters using all the fusion detection tools provided (arriba, fusioncatcher, pizzly, squid, starfusion and stringtie). Only fusions detected by at least two tools were considered as confident.
Xenium
Four patient FFPE tissues (ecDNA+ n = 2 and ecDNA− n = 2) were characterized with Xenium (10X Genomics) in situ spatial transcriptomics using the Human Multi-tissue and Cancer panel (377 genes) plus a custom panel of 37 genes (Supplementary Table 6). FFPE samples were processed and analysed according to the manufacturer’s protocol with no modifications. Post-Xenium H&E staining was performed as described in the Xenium protocols. We obtained a total of 807,253 cells, with a mean of 141.65 decoded transcripts for 100 µm2, and good quality of transcripts (Phred quality score ≥ 20) above 92%. After quality control, we retained 805,966 cells for subsequent analysis. Raw Xenium data were imported in Seurat (v5.1.0) and integrated using reciprocal principal component analysis to remove batch effect correction. Cell clusters were identified with Leiden clustering at a resolution of 0.2, and cluster markers were identified with R package presto (v1.0.0). Seurat cluster annotation was imported in Xenium Explorer (10X Genomics, v3.1.0) for visualization and integration with the H&E image.
Statistical analyses
All statistical analyses were carried out using R (v4.1.2) or GraphPadPrism (v9.5.1). A Fisher’s exact test and Chi-squared test were used to evaluate the significance in contingency tables. The Wilcoxon rank-sum test was used in two-group comparisons, and the relationship between two quantitative variables was measured using the Pearson correlation. Other statistical tests performed are described in the figures or in the figure legends.
Public datasets
Amplicon information for the ICGC PACA-CA and PACA-AU WGS samples was obtained from Kim et al.21. Additional matching ploidy data were retrieved from the ICGC Data Portal (https://dcc.icgc.org/releases/PCAWG). To focus on PDAC specifically, only PDAC tumours with histological types ‘8500/3’, ‘8560/3’, ‘8140/3’, ‘adenosquamous carcinoma’ and ‘pancreatic ductal adenocarcinoma’ were used in the downstream analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.