Mice
All animal procedures were conducted in accordance with a protocol approved by the National Institutes of Health Institutional Animal Care and Use Committee (IACUC), Bethesda, MD, USA, and complied with Public Health Service Policy on Humane care and Use of Laboratory Animals and the Guide for the Care and Use of Laboratory Animals. We used the following transgenic mouse lines: Emx1-IRES-Cre (JAX 005628)61, tetO-GCaMP6s (JAX 024742)62, and CaMK2a-tTA (JAX 007004)63. We performed experiments on 10 Emx1-IRES-Cre, 71 CaMK2a-tTA;tetO-GCaMP6s, and 3 GCaMP6s+.CaMK2a-tTA− mice. We used both male and female mice (females, 45%), 12–24 weeks old at the experimental endpoint. The percentage of Movup and Movdown neurons and sensory-responsive neurons was similar between the male and female groups (Movup and Movdown neurons, 30 ± 11% versus 33 ± 11%, n = 26 versus 21 mice, respectively, P = 0.27; sensory-responsive neurons, 13 ± 5.9% versus 12 ± 6.1%, n = 19 versus 14 mice, respectively; P = 0.57, two-sided Wilcoxon rank-sum test). Mice were housed in groups, in individually ventilated and enriched laboratory cages, in climate-controlled rooms (22 °C; 45% humidity), under a reverse 12 h light:12 h dark cycle (light on, 09:00), and with ad libitum access to water and food. After surgical procedures, mice were housed individually. All experiments were performed in the dark phase of the cycle. Mice in test and control groups were littermates and randomly selected.
Surgeries
All surgical procedures were performed stereotaxically, including injection of recombinant adeno-associated viruses (rAAVs), head plate implantation, and cranial window implantation, and were carried out under aseptic conditions. Mice were anaesthetized with isoflurane (1.0–2.0% in O2 at 0.8 l min−1). The eyes were protected with ophthalmological ointment, and body temperature was maintained at ~37 °C using a heating pad (Stoelting). Dexamethasone (0.2 mg kg−1 of body weight; subcutaneous injection) was administered at least 1 h prior to cranial window implantation, to prevent brain oedema. Exposed dura mater was perfused with sterile Ringer’s solution (in mM, 150 NaCl, 2.5 KCl, 10 HEPES, 2 CaCl2, 1 MgCl2; pH 7.3 adjusted with NaOH; 300 mOsm). After surgery, mice were treated with meloxicam (2 mg kg−1; subcutaneous injection) every 24 h for 3 days, to minimize pain and inflammation, and with enrofloxacin (0.1 mg ml−1 in drinking water) for 5 to 10 days, to prevent infection. Wellness and body weight were monitored daily for 10 days.
The first surgery consisted of rAAV injection combined with headpost and cranial window implantations, rAAV injection followed by headpost implantation, or headpost implantation only. For rAAV injection, at each target coordinate, the skull was thinned, and a craniotomy (~50 µm diameter) was made using fine forceps. A glass micropipette (5–10 µm outer diameter tip) attached to a nanoinjector (WPI) was used to deliver the viral vector (at 20–50 nl min−1). After injection, the pipette was left in place for ~5 min before being slowly retracted. To express GCaMP6f in PNs, we injected rAAV5-Syn-Flex-GCaMP6f-WPRE-SV40 (Addgene 100833) in the right hemisphere wS1 of Emx1-IRES-Cre mice (in mm relative to Bregma: anterior–posterior (AP) –0.80 and medial–lateral (ML) 3.50; AP –1.20 and ML 3.40, the pipette was angled at 21°, and 30 nl were injected at the subdural depths of 350, 250 and 150 µm). For GRAB sensor experiments, we injected AAV9-hsyn-Ach4.3 (Ach3.0) or AAV9-hsyn-NE2m (NE3.1) (WZ Biosciences) in the right hemisphere wS1 of GCaMP6s+.CaMK2a-tTA− mice (3 injection sites; same injection parameters as above). For simultaneous imaging of L2/3 PNs and optogenetic suppression of thalamic or motor cortical terminals in wS1, we injected rAAV5-CAG-ArchT-tdTomato (UNC Vector Core AV4595B), rAAV5-CAG-tdTomato (Addgene 59462), or rAAV5-Syn-tdTomato (Addgene 51506) in the VPm (AP −1.70, ML 1.85 and dorsal–ventral (DV) 3.15; 50–60 nl) and/or POm (AP 2.00, ML 1.32, and DV 3.00; 50–60 nl), or the M1/2 (AP −1.00, ML 1.00, and DV 0.8 to 0.2, 15 nl per each 100 µm) of tetO-GCaMP6s;CaMK2a-tTA mice. We implanted a custom-made Y-shaped titanium head plate using dental cement (Super-Bond C&B, Parkell). The exposed skull was covered with a thin layer of clear dental cement and, subsequently, opaque biocompatible silicone (Kwik-Cast, WPI), if applicable. A craniotomy was made over the right hemisphere wS1 (centred at AP 1.1 and ML 3.3), and a glass cranial window (diameter, 3 mm; thickness, 100–150 µm) was placed and secured over the craniotomy using cyanoacrylate adhesive (3M) and dental cement. For in vivo neuropharmacological experiments and single-neuron monosynaptic input tracing, the cranial window had a rectangular laser-cut opening (0.30 × 0.80 mm, Potomac Photonics) covered with transparent biocompatible silicone (Kwik-Sil, WPI)64. On the day of imaging, the silicone plug was removed and micro-durotomy was performed for direct access to the brain.
Recordings were initiated after a minimum period of 3–5 weeks post-rAAV injection, for stable expression of GCaMP6f, ArchT, GRABACh or GRABNE.
Behaviour
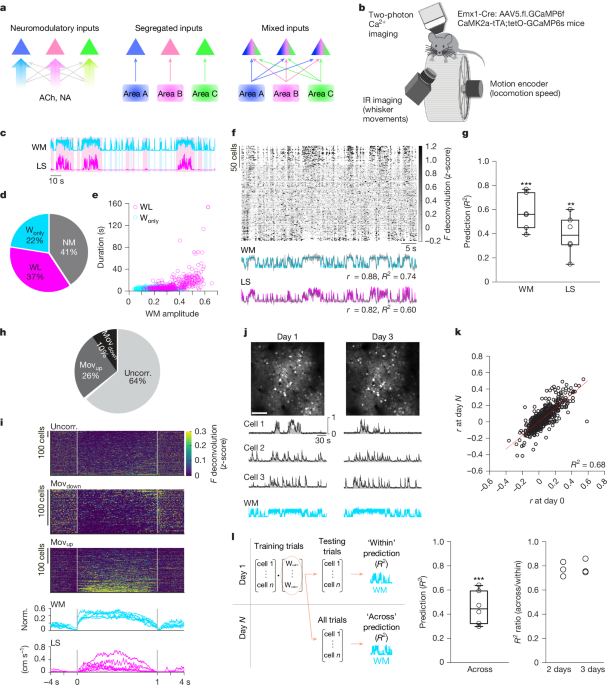
All behavioural experiments were performed in darkness. Under head-fixation, mice with all intact whiskers were free to run on a wheel. The mouse face and whiskers were video-recorded at 250 fps using a high-speed camera (acA2000-340kmNIR, Basler) with an 8 mm lens (LM8JC, Kowa), under infrared LED illumination (850 nm, Mightex). Image acquisition was controlled by StreamPix (NorPix). The wheel (diameter, 15 cm; width; 5.5 cm) was set so that only forward movement was permitted. To extract locomotion speed, we used a 2500 CPR resolution motion encoder (Model 260 Accu-Coder, Encoder) affixed to the wheel shaft. Motion encoder pulses were converted to speed using a counter and LabView software (National Instruments), for online visualization of speed. To synchronize behavioural and neuronal data, voltage signals from each video-frame exposure and wheel speed were digitized and recorded at 10 kHz through a data acquisition card (PCI-6052E, National Instruments), using the Prairie View Interface (Bruker).
Unilateral mystacial pad paralysis
In a subset of mice, paralysis of the left or both mystacial pad(s) was achieved through a local, subcutaneous injection of BTX (single injection per pad; 0.5 Units in 10 µl per injection, BOTOX), under isoflurane anaesthesia.
Whisker stimulation
Deflection of the left whiskers was achieved using a solenoid valve-controlled pole (diameter, 3 mm; length, 5 cm). To maximize contact with all whiskers, the pole was positioned in alignment with the mystacial pad, at an angle of ~65 ° (distance between the mystacial pad and the pole during stimulation, ~5 mm). Stimuli (trains of 28–49 stimuli; speed, ~600 mm s−1; duration, 50 or 250 ms; interval, 3–5 s) were produced and synchronized to behavioural and neuronal data using the Prairie View Interface (Bruker). To ensure that changes in neuronal activity related to whisker deflection could be isolated from changes in neuronal activity related to potential alterations in movements during deflection, recordings included trials of whisker deflections coupled with sound, as well as sound-only trials61. Neurons were classified as sensory stimulus-responsive if they responded exclusively during whisker deflections, but not during sound-only trials. The percentage of sensory-responsive neurons did not differ when using 50-ms-duration versus 250-ms-duration whisker stimuli (14 ± 5.9% versus 11 ± 6.0%, n = 21 versus 12 mice, respectively; P = 0.15, two-sided Wilcoxon rank-sum test).
In vivo imaging and optogenetics
Imaging was performed using a two-photon microscope (Ultima Investigator, Bruker) and a fs-pulse Ti:Sapphire laser (Mai Tai DeepSee, Spectra-Physics), tuned between 860 and 1040 nm, for imaging of difference fluorescent proteins. The microscope was equipped with an 8 kHz resonant galvanometer and a water-immersion 16× objective (0.8 NA, Nikon) coupled to a 400-µm-range, z-axis piezoelectric drive. GCaMP6 and RFP fluorescence signals were passed through a 525/70 m or 595/50 m filter, respectively. Fluorescence was detected and amplified using GaAsP PMTs (Hamamatsu) and a dual preamplifier, prior to digitization. For two-photon Ca2+ imaging, we performed one session per day (recording time ~66 ± 21 min). Images (resolution, 512 × 512) were collected at ~30 Hz, in single plane mode. FOVs ranged from 271 × 271 µm (for functional identification of a neuron for subsequent electroporation) to 573 × 573 µm (for characterization of neuronal patterns of activity) with an average excitation laser power of ~33–76 mW at the objective. For simultaneous two-photon Ca2+ imaging and optogenetic inhibition of thalamic axon terminals, we used a collimated 625 nm LED beam (Prizmatix UHP-T-625-SR)65,66. The LED was on during the turnaround of the resonant galvanometer. Trains of light pulses (25 pulses; duration per pulse, 1–1.5 s; interval, 5 s) were generated and synchronized to behavioural and neuronal data acquisition using the Prairie View Interface (Bruker). A filter (FF02-617/73-25, Semrock) was used to narrow the LED spectrum, and a dichroic mirror (FF556-SDi01, Semrock) was used direct the LED light onto the brain tissue and pass GCaMP6 florescence signals onto the PMT. The average power of the LED was 10–50 mW at the objective. To minimize the effect of light stimulation on the spontaneous movements of mice, we shielded the objective lens. However, this shield did not completely block the light stimulation. We observed a brief (<0.5 s) whisker movement in both control and ArchT-expressing mice at the onset of light stimulation (Extended Data Fig. 16).
In vivo neuropharmacology
A durotomy (∼50 µm) was made through the access port of the implanted cranial window, under isoflurane anaesthesia (Fig. 2a). Mice were allowed to recover for ∼30 min prior to recordings. Following acquisition of baseline behavioural and neuronal activity data, Ringer’s solution was replaced by Ringer’s solution supplemented with receptor blockers, and recordings were reinstated. In sham sessions, Ringer’s solution was replaced by Ringer’s solution without blocker addition. Only data acquired 20 min or more after blocker application or Ringer’s replacement (in sham sessions) were considered for analysis. Each session, 1–2 days apart, lasted a median of 2 h 15 min (effective spontaneous activity recording time, 1 h and 8 min): baseline period, 42 min (effective spontaneous activity recording time, 30 min); blocker application or Ringer’s replacement, 3 min; waiting period following blocker application or Ringer’s replacement, 21 min; receptor blockade period, 45 min (effective spontaneous activity recording time, 30 min). Baseline versus sham/receptor blockade recording times did not differ (P > 0.05, two-sided Wilcoxon signed-rank test). We used a combination of atropine (1 mM) and mecamylamine (1 mM) to block ACh receptor (AChR)37,38,67,68, a combination of prazosin (1 mM) and propranolol (1 mM) to block noradrenaline receptor (NAR)69, d-AP5 (1 mM) to block NMDA receptor (NMDAR), and a combination of d-AP5 and DNQX (2 mM) to block both NMDA and AMPA receptor (AMPAR). In one mouse, we used prazosin (1 mM), propranolol (1 mM) and yohimbine (1 mM) to block the noradrenaline receptor. Blocker application session sequences were either AChR–NAR–NMDAR/AMPAR (n = 3) or NAR–NMDAR/AMPAR–AChR (n = 2), randomly assigned per animal. In two mice, only ACh receptor (n = 1) or noradrenaline receptor (n = 1) blocker sessions were performed. The position of ACh or noradrenaline receptor in the sequence did not affect neuronal correlations (P > 0.05, linear mixed model controlling for days as confounding factor for position). To test the effectiveness of drug diffusion into the imaging FOV (481.4 × 481.4 to 572.9 × 572.9 µm2, at 330 ± 30 µm of depth), we applied TTX (10–100 µM) in the final recording session. Experiments in which TTX did not silence neuronal activity over the entire FOV within 15–20 min were excluded (n = 2 mice).
To evaluate the effectiveness of ACh and noradrenaline receptor blockade throughout the entire FOV, we performed equivalent neuropharmacological experiments in mice expressing either GRABACh or GRABNE in wS1 L2/3 neurons.
In vivo single-neuron monosynaptic input tracing
After micro-durotomy, we performed two-photon Ca2+ imaging and selected a target neuron based on its activity profile across behavioural states. Classification of the target neuron as movement-uncorrelated or movement-correlated was confirmed during post-hoc analysis. After imaging, the mouse was lightly anesthetized, and two-photon guided electroporation of the target neuron was performed as described previously, for monosynaptic input tracing14,17,39,70,71,72. A glass pipette (14 ± 1.5 MΩ) was filled with intracellular solution (in mM, 130 potassium gluconate, 6.3 KCl, 0.5 EGTA, 10 HEPES, 5 sodium phosphocreatine, 4 Mg-ATP, 0.3 Na-GTP; pH 7.4 adjusted with KOH; 280–300 mOsm) supplemented with Alexa 594 hydrazide (50 µM, A10442, Thermo Fisher Scientific) and two DNA plasmids (pAAV-EF1α-mTagBFP-HA-T2A-mCherry-TVA-E2A-N2c, 0.15 µg µl−1; pAAV-CAG-N2c, 0.05 µg µl−1). The resistance of the pipette tip was monitored continuously (Axoporator 800A, Molecular Devices). Positive pressure was applied to the pipette (70 mbar), which was visually advanced through the durotomy, using a micromanipulator (PatchStar, Scientifica). Upon entering the cortex, the pressure was swiftly decreased (35 mbar). Then, within ~50–100 µm from the target neuron the pressure was further decreased (15 mbar). The pipette was slowly advanced towards the soma of the target neuron, until the tip resistance increased by at least 20%. The pressure was released, and a train (100 Hz, 1 s) of electric pulses (−10 V, 0.5 ms) was applied (Axoporator 800A), after which the pipette was retracted. The electroporated neuron was imaged 20 min later to evaluate its survival. Thereafter, we injected G-deleted, envelope-A coated CVS-N2c rabies virus carrying RFP (kindly provided by the Center for Neuroanatomy with Neurotropic Viruses) in the vicinity (within ~150 µm) the electroporated neuron (rate, 30 nl min−1)73. Then, the access port of the cranial window was sealed using biocompatible silicone. Survival and successful transfection of the electroporated neuron was monitored within 2–3 days after electroporation and up to the experimental endpoint. Structural, two-photon z-stacks (1–5-µm steps; resolution, 512 × 512; FOV, 102 × 102 to 271 × 271 µm) including the imaging FOV and/or the target neuron were acquired before and after electroporation, to track individual cells volumetrically throughout the experiment. Local, wS1 presynaptic networks were followed structurally through two-photon imaging of GCaMP6 and RFP (z-stacks, 1–5-µm steps; resolution, 512 × 512; FOV, 271 × 271 to 814 × 814 µm). For z-stack acquisition the average laser power was depth-adjusted linearly and did not exceed 100–150 mW at the objective. Mice were euthanized at day 11 (±1.5 days) following electroporation, and brains were processed for ex vivo input tracing. Brains containing less than 100 presynaptic cells were excluded from analysis (n = 1).
Histology
Upon completion of recordings, mice were deeply anesthetized and perfused transcardially with 4% formaldehyde in PBS. Post-perfusion, brains were immersion-fixed in 4% formaldehyde in PBS for 2–3 h and then transferred to 30% sucrose in PBS.
For input tracing experiments, whole-brain free-floating sequential coronal sections (50-µm-thick) were obtained using a microtome (SM2010R, Leica). Sections were rinsed 3 times in PBS, incubated in blocking solution (5% normal serum and 1% Triton X-100 in PBS) at room temperature for 1 h, and subsequently incubated in primary antibody solution (2% normal serum and 1% Triton X-100 in PBS) at 4 °C for 48 h. Primary antibodies were detected through incubation in secondary antibody solution (2% normal serum and 1% Triton X-100 in PBS) at room temperature for 2 h. We used the following primary and secondary antibodies and respective dilutions: anti-RFP 1:500 (600-901-379, Rockland); anti-GABA 1:500 (A2052, Sigma), IgY-Alexa Fluor 555 1:200 (A21437, Thermo Fisher Scientific); IgG-Alexa Fluor 647 1:200 (A21245, Thermo Fisher Scientific). Sections were rinsed in PBS and sequentially mounted on glass slides. Neuronal nuclei were revealed through a fluorescent Nissl stain (NeuroTrace 435/455, N21479, Thermo Fisher Scientific), after which sections were cover-slipped. Whole-brain serial sections were imaged using an epifluorescence illumination microscope (Zeiss Imager.M2). Multiple z-stacks (10 µm steps), covering each section in its entirety, were acquired using Neurolucida (MBF Bioscience). z-stacks were aligned and collapsed onto a single image using Deep Focus (Neurolucida).
For all other experiments, brain sections were similarly generated and mounted, and neuronal nuclei were visualized either using fluorescent Nissl stain (NeuroTrace 435/455 or NeuroTrace 530/615, N21482, Thermo Fisher Scientific) or DAPI (Fluoromount-G mounting medium, Thermo Fisher Scientific). Entire sections were imaged using an Axio1 Scanner (Zeiss). FOV location within wS1 was confirmed either by targeted two-photon laser microlesions (the laser beam was focused at a subdural depth of 200 µm; 800 nm; 5–30 s; ~0.5 W)74 or fluorescent dye (DiI (42364, Sigma) or Fast Blue (17740, Polysciences)) injection at the experimental endpoint and ex vivo histological analysis. Tissue imaging and histological analysis were done blinded to the experimental groups. For analysis of thalamic ArchT-expression areas, a composite image of a brain section at the injection centre for either VPm or POm was selected, and expression areas annotated; each brain section was manually aligned to the corresponding mouse brain atlas section, and the expression areas of the different mice were overlaid75,76,77.
Defining behavioural events
Video recordings of face and whiskers were processed using a custom MATLAB routine. To detect whisker movements, we first defined a region of interest (ROI) encompassing the left or right whiskers in an image that consisted of the s.d. of representative frames of each session. We then computed the absolute power of the spatial derivative of consecutive frames (WM trace). Both the whisker movement and locomotion speed traces were downsampled to 30 Hz, averaging the values acquired during a Ca2+ imaging frame. To detect behavioural events, the whisker movement trace was baseline-subtracted (10th percentile of the full trace) and normalized. We then applied a threshold to the whisker movement trace (3× minimum s.d., calculated using a 30-s sliding window). Detection was visually inspected for all sessions, and the threshold was adjusted when applicable. Then, local maxima were calculated, and peaks less than ~0.5 s apart were considered as part of a single event; the first peak was considered as onset and the last, as offset. Events with an integral value smaller than 20 (<3% of the session time) were excluded from analysis. Whisker movement events were considered as WL when the maximum locomotion speed was higher than 0.20 cm s−1 and, conversely, as Wonly when the locomotion speed did not exceed 0.20 cm s−1. For comparisons across mice, the raw whisker movement trace was baseline-subtracted and normalized to its maximum; for BTX experiments, across session data were normalized to maximum according to the first session.
Processing of two-photon calcium images
Two-photon Ca2+ images were processed using Suite2p78, in Python, with default parameters, unless otherwise indicated. Following subtraction of neuropil (fixed scaling factor of 0.7) and baseline (calculated on filtered traces, using a gaussian kernel of width 20 and a sliding window of 60 s), fluorescence traces were deconvolved using non-negative spike deconvolution79 with a fixed decay timescale of 0.7 s for GCaMP6f and 1.5 s for GCaMP6s. To ensure that only somatic traces were included in the analysis, ROIs were manually curated by an analysist blinded to the experimental group. Aligned image series were visually inspected to control for z-drifts; data showing z-drifts were excluded from analysis. Tracking of the same neurons across sessions was done semiautomatically using a MATLAB script. All analysis was based on deconvolved traces; for presentation purposes only, we used neuropil subtracted fluorescence traces normalized to the maximum (F), overlaid with deconvolved fluorescence traces normalized to the maximum (F deconv.), unless otherwise indicated. To generate temporal raster plots (Fig. 1e), the activity of each neuron was averaged over ~0.5 bins, z-scored and smoothed using a 1-s moving average filter; individual neurons were sorted by the first principal component of neuronal activity.
Processing of two-photon GRAB sensor images
Two-photon Ca2+ images were motion-corrected using Suite2p78. Thereafter, we extracted the mean fluorescence intensity of pixels within 6 ROIs (75 × 75 pixels) manually spread over the entire FOV, avoiding large vessels. In addition, to extract a full FOV fluorescence intensity mean, we first isolated sensor+ pixels and excluded vessel-related pixels by applying a threshold to pixel intensity (99.9th percentile > intensity > 50th percentile) over the motion-corrected, whole recording session average image80. Baseline traces were obtained essentially as described in ‘Processing of two-photon Ca2+ images’ but using a gaussian kernel of width 30 and a sliding window of ~120 s. The mean fluorescent values of single ROIs or full FOVs were baseline-subtracted (ΔF) and z-scored.
Decoding analysis
We trained a linear decoder to decode behavioural variables from neuronal activity. We minimized the ridge regression81 objective function (equation (1)).
$$\widehat{w}=\mathop{{\rm{argmin}}}\limits_{w}\mathop{\sum }\limits_{i=1}^{N}{\parallel {y}_{i}-{w}^{T}{x}_{i}\parallel }_{2}^{2}+{\rm{\alpha }}{\parallel w\parallel }_{2}^{2}$$
(1)
where yi is the behavioural variable at time (frame) i, xi is the neuronal activity matrix, w is the weight vector, and α is the ridge parameter (regularization). We normalized the behaviour and neuronal activity by z-score for the cross-day recordings. We did not normalize the neural activity for the neuromodulatory experiments, as activity was recorded continuously on the same day. For optogenetic experiments, we ran analysis with (shown in figures) and without data normalization, as well as with and without (shown in the figures) rebound cells, and no substantial difference was found. We randomly split the trials into training (75%) and test sets (25%). The weight vector was estimated on the training set, and the ridge parameter was selected by leave-one-out cross-validation82 on the training set.
We evaluated the decoding performance by the out-of-sample (test set) coefficient of determination (R2) (equation (2))81.
$${R}^{2}=1-\frac{{\sum }_{(i)}\,{({y}_{(i)}-{\hat{y}}_{(i)})}^{2}}{{\sum }_{i}\,(\,{y}_{(i)}-{\bar{{\rm{y}}}}_{(i)}{)}^{2}}$$
(2)
where (i) is the index of out-of-sample trials, \({\widehat{y}}_{(i)}={\widehat{w}}^{T}{x}_{(i)}\) and \(\widehat{w}\) is the estimated weight vector from the training set by (1). Using the weight vector estimated from the training set, we decoded the behavioural variables on the test (held out) set within the same condition or session (referred as within) and the trials of other conditions or sessions (referred as across). Then, we calculated the out-of-sample R2 using the predicted and true values for within and across data. The out-of-sample R2 is not prone to overfitting and will not be inflated. Note that the out-of-sample (test set) R2 can be negative if the fitted model does not predict the test set at all, which indicates that the neural correlations could be substantially different between the training and test sets.
Modulation of neuronal activity
Data were analysed using MATLAB scripts. The activity (F deconvolved) of each neuron was aligned to the onset and offset of spontaneous movements, Wonly and WL. Baseline and post-offset activities refer to a 0.5-s window preceding movement onset and a 0.5-s window after movement offset, respectively. A neuron was considered as Movup if its average activity during Wonly and/or WL events was significantly higher than its average activity during baseline and/or significantly higher than its average activity post-event (P < 0.01, two-sided paired-sample t-test). Conversely, a neuron was considered Movdown if its average activity during Wonly and/or WL events was significantly lower than its baseline and/or significantly lower than its post-event average activity (P < 0.01, two-sided paired-sample t-test). Neurons exhibiting opposite changes in activity for Wonly and WL were rare (0.7 ± 0.2% of all cells) and were not considered for further analysis. Otherwise, neurons were considered as movement-uncorrelated. Modulation refers to the mean activity during movement, irrespective of movement duration, minus the mean activity during baseline, averaged across spontaneous movements (Wonly, WL, or Wonly + WL).
Time-normalized PETHs were created by normalizing the data to match the average duration of WL events across mice. To compute the correlation (Pearson’s linear correlation coefficient, r) between the activity of individual neurons and each behavioural variable, deconvolved fluorescent traces, and corresponding whisker movements and locomotion speed raw traces were binned (bin size, ~0.5 s).
Sensory stimulus-responsive neurons were defined by a significantly higher average activity within a response window of 0.5 s after the onset of whisker stimulation (coupled with sound) versus baseline (P < 0.01, two-sided paired-sample t-test). We excluded cells that also responded to sound-only stimuli versus baseline (P < 0.01, two-sided paired-sample t-test). Baseline was calculated on a 0.5-s window preceding stimulus onset. Sensory stimulus-response magnitude refers to the mean activity during the response window minus the mean activity during baseline, averaged across all whisker stimulations.
For neuropharmacological experiments, the distribution of modulation values for each WL neuron in the presence of receptor blocker(s) was compared to that of baseline (prior to blocker application, two-sided t-test). Similarly, we compared the distribution of sensory stimulus-response magnitude values before and after blocker application for each sensory stimulus-responsive neuron (two-sided t-test).
For optogenetic experiments, light-off periods were used to identify neurons as movement-uncorrelated or movement-correlated. Neurons were classified as light modulated if their activity during the light pulse differed significantly from that of baseline (0.5-s window prior to light pulse onset). Most L2/3 PNs fire sparsely; only neurons that showed an average baseline activity (F deconvolv.)—that is, prior to the light pulse, higher than the 75th percentile of the average baseline activity across all cells were included in the analysis. To generate time-normalized PETHs, light pulse data were normalized to match the maximum pulse duration across experiments (1.5 s). Statistical testing on PETHs of movement-uncorrelated and movement-correlated neuronal subpopulations was performed on the original data sampling rate (~30 Hz).
Whole-brain reconstruction, annotation and registration
To analyse the brain-wide distribution of presynaptic neurons, we adapted a previous pipeline83,84. To reconstruct a whole-brain 3-dimensionally, individual section images were aligned using BrainMaker (MBF Bioscience). Brain-wide presynaptic neurons (RFP+) were automatically segmented using NeuroInfo (MBF Bioscience) and manually annotated according to brain area. Local, wS1 presynaptic neurons were manually annotated based on cortical layer location. Distinct layers were identified based on the characteristic depth-varying density of NeuroTrace+ neurons. Presynaptic neurons within each layer were identified as glutamatergic (GABA−) or GABAergic (GABA+). To confirm the colocalization of RFP and GABA, a subset of brains was re-imaged using a confocal microscope (z-stacks, 3-µm steps, C2, Nikon). Identification of glutamatergic and GABAergic neurons was equivalent for the two different imaging methods. Each serially reconstructed brain was registered to the Allen Mouse Common Coordinate Framework, and brain-wide presynaptic neurons (RFP+) were automatically re-identified according to distinct anatomical structures. Reconstructions and registrations were conducted blindly. Manual identification was independently performed by two analysts (one analysist was blinded to the activity profiles of the postsynaptic neurons). All manual and automatic identifications were coherent.
Analysis of brain-wide presynaptic networks
Fine-scale spatial registration of wS1 presynaptic networks
Postsynaptic neurons did not survive until the experimental endpoint14. We used the centre of mass of glutamatergic presynaptic networks in L2/3 to estimate the position of the postsynaptic neurons for all 22 subjects14, which were then averaged to construct a reference postsynaptic site on the Allen Mouse Common Coordinate Framework. From the brain atlas, we manually marked the boundaries of the cortical surface and performed surface triangulation. Using this triangulated cortical surface, we then estimated the surface’s normal vector that goes through the reference postsynaptic site and serves as the reference normal vector. Individual wS1 presynaptic network of each subject was then rigidly aligned based on the position of the reference postsynaptic site and orientation of the reference normal vector.
Layer-by-layer horizontal flat projections
After the fine-scale registration, we concatenated all neurons from both movement-uncorrelated and movement-correlated groups. Then we performed PCA on each layer of the presynaptic networks to obtain the population best-fitted plane. Corresponding neurons from the layer were then projected onto the best-fitted plane. We then performed a rigid parameterization and mapped the neurons to a two-dimensional coordinate system. The resulting parameterization of each layer from every subject with gaussian kernel density estimation is visualized in Extended Data Fig. 11.
Statistical analysis of group-wise spatial distribution differences
To explore whether there was any difference in the spatial pattern of presynaptic networks between movement-uncorrelated and movement-correlated groups, we tested the null hypothesis of no spatial distribution difference between multiple local and long-range anatomically annotated presynaptic neurons of the two groups. The final statistical analysis incorporated Bonferroni correction. We chose the 2-Wasserstein distance function as the test statistics, and we performed a one-sided randomization test (n = 10,000) to approximate the permutation distribution85,86. Owing to the variability of total number of presynaptic neurons across brains, we introduced non-uniform sample weights when we computed the 2-Wasserstein distance to avoid any dominated effect from subjects having a large number of presynaptic neurons. Specifically, we first re-weighted every neuron by the inverse of the number of (for example, layer-wise/whole-wise) presynaptic neurons to ensure an equivalent contribution from subjects with non-empty neuron sets. Second, sample weights from subjects in the same group were concatenated and normalized into a probabilistic mass. This alleviates the imbalance of group-wise total mass if some of the subjects have no neurons detected in specific cortical layers (for instance, GABAergic presynaptic neurons in layer 6). To aid in the visualization of the spatial spread of presynaptic networks across cortical areas, we generated cortical flat maps. The 3D Allen Common Coordinate Framework coordinate points of registered neurons were projected along streamline paths orthogonal to the cortical surface to create a flat map 2D representation that preserves relative spatial position of cortical areas for analysis using tools provided by the Allen Institute87.
Statistical analysis of group-wise proportions of M1 and M2 presynaptic cells
Motor cortical neurons that project to wS1 are distributed closely along the anatomical border between M1 and M288. To evaluate whether the movement-uncorrelated and movement-correlated groups exhibited an M1 or M2 bias in presynaptic cell proportions, in addition to the fraction of M1 and M2 cells, we computed the shortest 3D Euclidean distance of each M1/2 cell from the anatomical border between M1 and M2 (represented by an open surface mesh in the Allen Mouse Common Coordinate Framework). Distances were assigned with negative or positive values based on whether cells were in M1 or M2, respectively. Given the variability in cell counts across subjects, we implemented a reweighting procedure on the distance distribution, aiming at an equitable contribution of each subject to the group. This involved resampling cells based on a probability assigned to each cell, inversely proportional to the cell count within each subject, resulting in group-wise distribution of distances based on 10,000 resampled cells. Owing to a minimal number of M1/2 cells, four mice in the movement-uncorrelated and four mice in the movement-correlated groups were excluded. The observed mean difference was compared to a distribution of mean differences obtained by randomly permuting the group labels and recalculating the weighted mean difference for each permutation. This process was repeated 5,000 times to obtain the permutation distribution.
Statistics
We did not use statistical methods to predetermine sample size. All data were acquired according to standard protocols, batch processed using the same codes, and independently processed and analysed by analysts blinded to the experimental groups. Analytical routines and were established using a subset of the data (training set), and these were applied to entire datasets, whenever applicable to avoid overfitting. Data were presented as mean ± s.d. throughout the text. In box plots, the central line represents the median, the box represents the 25th and 75th percentiles, and the whiskers extend to the most extreme data points excluding outliers (larger than 1.5× the interquartile range); when overlaid with individual data points, all data points, including outliers, were graphed. Statistical tests used were indicated in the figure legends. All comparisons using two-sample or paired-sample t-tests, Wilcoxon rank-sum or signed-rank tests were two-sided unless otherwise indicated. Linear fixed effects models (with interaction between drug type and neuronal correlations) were used to compare slopes (Fig. 2). Linear mixed effects models were used to test effect of receptor blocker application order. Bonferroni correction was applied to multiple comparisons unless otherwise indicated. Significance levels are indicated as: NS, not significant (P ≥ 0.05); *P < 0.05; **P < 0.01; ***P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.