Human subjects
Adult male and female patients at Massachusetts General Hospital and Brigham and Women’s Hospital (MGB) provided preoperative informed consent to take part in the study in all cases under the approved Institutional Review Board Protocol DF/HCC 10-417. Patients’ clinical characteristics are summarized in Supplementary Table 1. Patients in other cohorts gave consent according to published methods7,10,13. Previously unpublished patient data from the Montreal Neurological Institute was collected as reported with other tumours from McGill University23.
Primary tumour processing for Seq-Well and GBOs
Fresh tumour samples were collected directly from the operating room at the time of surgery and the presence of glioma was confirmed by frozen section. Samples were dissected into small pieces and mixed. For samples with enough material, we divided up the mixed tumour pieces, with part being used for single-cell dissociation and part for GBO generation.
Single-cell dissociation and Seq-Well
For the MGB cohort, minced tissue pieces were mechanically and enzymatically dissociated using a tumour dissociation kit, according to the manufacturer’s instructions, and a GentleMACS Octo Dissociator with Heaters (Miltenyi Biotec) using custom settings. The single-cell suspension was then depleted of dead cells and debris using magnetic-activated cell sorting (Dead Cell Depletion Kit, Miltenyi Biotec). Cells were then processed for Seq-Well as previously described56. In brief, cells were resuspended in media and 10,000–15,000 cells were then distributed dropwise onto a Seq-Well microwell array preloaded with mRNA capture beads. The arrays were then sealed with a semipermeable membrane and submerged in lysis buffer to lyse their cellular and nuclear membranes. Lysis buffer was then removed and the array was submerged in a hybridization buffer to allow for attachment of mRNA to the capture beads. After mRNA capture, the seal was removed from the array and the capture beads were manually washed out of the array with a washing buffer. Following removal from the array, the beads were washed to remove any excess lysis reagents and unattached nucleic acids. A reverse-transcription reaction was run on the beads and cDNA was generated by elongation of the capture bead oligos using the captured RNA as a template. An exonuclease reaction was then run on the beads to clean them up. After this, a second strand-synthesis reaction was run to generate a DNA strand complementary to the cDNA strand by polymerase chain reaction. Subsequent to second-strand generation, the now double-stranded cDNA was amplified by polymerase chain reaction in a whole-transcriptome amplification. Sequenceable libraries were generated from the whole-transcriptome amplification using the Nextera Library Prep kit, following the manufacturer’s instructions.
MAESTER materials and preparation
From the Seq-Well cDNA, mitochondrial RNA fragments were selectively pulled down, separately amplified and sequenced as previously described30.
Creation and maintenance of GBOs
Minced tissue pieces were further dissected using two scalpels until pieces were 1–2 mm in diameter. These were washed, further processed and maintained according to a previous protocol57.
Patient PBMC and monocyte processing
Patient PBMCs were collected at the time of surgery and isolated using SepMate-15 tubes (StemCell Technologies) and Lympholyte-H (Cedarlane) according to the manufacturers’ instructions. Cells were either directly processed for Seq-Well as above or enriched for pan-myeloid cells using CD11b beads (Miltenyi Biotec, 130-097-142) on Miltenyi magnet according to the manufacturing protocols, and then processed for Seq-Well. CD11b+D45+ purity was checked by flow cytometry (purity > 90%).
GBO perturbation and single-cell read-out methods
GBO perturbations
For perturbation experiments, GBOs were pipetted into ultralow-adherence round-bottomed 96-well plates (Corning, 7007) with one GBO per well. GBOs were plated in 100 μl GBO media. Small molecules were then added in an extra 100 μl of media at 2× concentration. Medium was changed every 2–3 days by removing 100 μl and replacing it with 100 μl fresh media with perturbation. Depending on the experiment, each condition had 6–12 GBOs per condition to account for heterogeneity among GBOs. For experiments with flow cytometry or scRNA-seq as a read-out, multiple GBOs were grouped together in replicates per condition and then dissociated to single cells together.
Myeloid–GBO co-culture
Human CD11b+CD45+ cells isolated from tumour or donor patient PBMCs, as described above, were aliquoted and frozen at 5 × 106–1 × 107 cells per ml per vial. Before co-culturing with GBOs, myeloid cells were gently thawed and washed in warm myeloid cell media (ImmunoCult-SF Macrophage Differentiation Medium, using base medium with only M-CSF (50 ng ml−1; STEMCELL Technologies, 10961), and plated in a 24-well low-attachment plate (Corning) to recover for 30 min in a 37 °C CO2 incubator. Plates were placed on an orbital rotator at 120 rpm with 2.5 × 106 maximum cells per well to avoid cell attachment and to maintain monocyte morphology. Monocytes (10,000–50,000, depending on the experiment) were then added to each GBO well in 100 μl myeloid cell media with small-molecule perturbation when applicable. Medium was changed every 2–3 days by removing 100 μl and replacing it with 100 μl fresh medium (a 1:1 mix of GBO medium and myeloid cell medium) with perturbation when applicable.
Dissociation of GBOs
In brief, all GBOs in each experimental replicate were grouped together in a 1.7 ml Eppendorf tube, the medium was aspirated and GBOs were washed twice with 1 ml media to remove small molecules and/or cells. GBOs were then dissociated to single cells using dissociation media from a Miltenyi tumour dissociation kit mixed 2:1 with accutase in the 1.7 ml tubes. These were placed at 37 °C and mechanically dissociated every 5–10 min by pipetting up and down until there was a homogeneous single-cell mixture. Cells were passed through a 40 μm filter and then used for downstream assays. Cells were processed for Seq-Well as described above or analysed by flow cytometry as described below.
Flow cytometry
Flow cytometry was based on a previous protocol58. In brief, an antibody cocktail was made by a 1:1 ratio of Brilliant Stain Buffer (BD Horizon, 566349) and PBS, and antibodies or dyes were added (antibodies are listed in Supplementary Table 5). Single-cell suspensions were washed by PBS + 0.5% BSA in 1.5-ml Eppendorf tubes or 96-well U-bottomed plates. Cells were pelleted by centrifugation at 300g for 5 min at room temperature. After removing the washing buffer, cells were resuspended in 100 μl staining cocktail by pipetting up and down about ten times. Plates or tubes were covered to avoid light and stained in the dark at room temperature for 20–25 min. Cells were washed by PBS + 0.5% BSA and centrifuged at 300g at room temperature for 5 min. Cell pellets were resuspended in 200 μl PBS + 0.5% BSA. Flow cytometry was processed on a BD LSRFortessa X-20 according to the manufacturer’s procedure. UltraComp eBeads (Invitrogen, 01-3333-42) were used to pre-annotate the compensation. FlowJo v.10 was used for data analysis.
Histological assessment of GBO experiments
GBOs were fixed in 4% formaldehyde for 30 min and then washed with DPBS and left in a 30% sucrose solution overnight to dehydrate the tissue. The organoids were then embedded in OCT (Sakura Tissue Tek) and frozen using an isopropanol bath. The tissue was then sectioned at a thickness of 8 μm using a cryostat. For staining, slides were dried at room temperature for 10 min and then prewashed with 1× TBST to remove the OCT. The tissue was blocked with a glycine BSA solution for 1 h at room temperature. Tissue sections were then incubated with primary antibodies either at 4 °C overnight or at room temperature for 2 h (antibodies are listed in Supplementary Table 5). Tissue sections were then washed thoroughly with 1× TBST and incubated with fluorophore-conjugated secondary antibodies and DAPI for 1 h at room temperature. The tissue was washed with 1× TBS and incubated with 1× True Black Autofluorescence quencher for 1 min. Tissue sections were washed with 1× TBS and mounted with Prolonged Gold mounting media (Invitrogen) and covered with glass slips. For imaging, a Leica Thunder microscopy system was used with an automated mechanized stage. Images were taken using the scanning features with a ×40 oil-immersion objective lens. Images were stitched together and enhanced with the fast computational clearing programs of Leica LAS X software v.3.8.2.27713.
Histological analysis
All histological images were analysed using Qupath open-source image-analysis software v.0.4.3. Cells were counted with the cell-detection feature using the DAPI channel. The detected cells were then called for positivity of up to three fluorescent markers using the single-object measurement feature with positivity thresholds adjusted according to the experiment. Thresholds were set by comparison of experimental conditions to control and then applied to all images of the experiment through automated scripts.
Single-cell, spatial and bulk RNA-seq analyses
Cohorts
This study used four cohorts, split into two datasets. The discovery dataset contained the MGB, Houston Methodist7 and Jackson Laboratories13 cohorts. Cells in these cohorts were assayed with more-advanced scRNA-seq technologies: Seq-Well S3 (MGB) or 10x Genomics 3′ v.3 (Methodist and Jackson Laboratories). The validation dataset was composed of samples from the McGill cohort, including a set that had been previously published10,23 and a set that was not previously published. McGill tumours were assayed using 10x Genomics 3′ v.2 kits. The Methodist data were downloaded (GEO series, GSE182109), the Jackson Labs data were downloaded from https://ega-archive.org/datasets/EGAD00001007772 and published McGill data were downloaded from https://ega-archive.org/datasets/EGAD00001006206.
For other tumours, we obtained raw expression matrices from sources indicated in Supplementary Table 4.
Alignment
We used the Cumulus platform59 to process the large-scale single-cell RNA-seq experiments. Libraries were aligned to the GRCh38 genome using STARsolo v.2.7.10 (ref. 60). Our GitHub page contains the specific settings (https://github.com/BernsteinLab/Myeloid-Glioma).
Data processing and visualization
The raw matrices outputs of STARsolo (no filtration of cells) for each tumour and GBO library were gzipped and used as input for Seurat61 using the Read10X() function with the default parameters. The pipeline was done for each cohort independently. Tumours in each cohort were merged using the Seurat merge() function to generate a Seurat object for each cohort. The percentage of mitochondrial gene expression was determined using PercentageFeatureSet() with the pattern set to ^MT. Cells with more than 25% of their transcriptome composed of mitochondrial gene expression were filtered out. We also filtered out cells expressing fewer than 500 genes or more than 6,000 genes. We also filtered out cells with less than 1,000 UMIs and genes expressed in less than three cells in the matrix. The filtering process was done using the Seurat subset() function.
For plotting, normalization, scaling and variable gene detection were done using the SCTransform() function, for which we used the percentage of mitochondrial gene expression as a regression factor. We performed principal components analysis (PCA) using RunPCA() with default parameters and generated an elbow plot using the ElbowPlot() function to determine the dimensions for generating UMAPs and for Louvain clustering (MGB, 24; Houston Methodist, 19; Jackson Laboratories, 16).
UMAPs were generated using RunUMAP() with the reduction set to pca. FindNeighbors() and FindClusters() were used for clustering, with the resolution set to 0.3.
Classification of cell types
To classify tumour cells in all cohorts, we identified the main cell programs in the MGB cohort and identified the top program for each cell in all cohorts. This top program was then used as the cell’s classification.
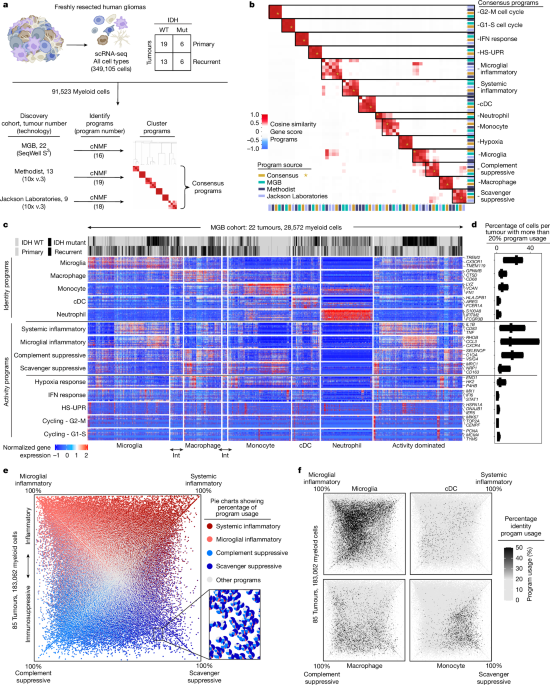
We merged all cells from the 22 tumours in the MGB cohort and used this expression matrix as the input for cNMF. We identified the 4,000 most variable genes using SCTransform, regressing out mitochondrial content. We subsetted the matrix for these genes and the resulting matrix was then subjected to consensus cNMF v.1.0.4 (ref. 24).
For the cNMF ‘prepare’ function, we performed factorization over K ranges from 2–35. We ensured that all the variable genes were considered for the factorization using the parameter –numgenes 4000. We also performed 500 iterations by inputting –n-iter 500 in the cNMF prepare script. K = 18 was the highest value with a silhouette score greater than 0.8 and was thus chosen for the consensus script of cNMF. cNMF was run with a –local-density-threshold value of 0.015.
We annotated each program on the final gene_spectra output of cNMF by comparing the top 100 genes with previously published gene sets and known marker genes. We used gProfiler62 to determine enrichment scores for a manually curated gene-set matrix with more than 600 gene sets (Supplementary Table 3). Manual integration of enrichment scores and known marker genes enabled us to determine the names of the programs (Extended Data Fig. 1a). Of note, MGH720, a tumour with a histological diagnosis of giant cell glioblastoma, had a cNMF unique malignant program.
We then used the gene spectra output of the cNMF programs to calculate the usage of these programs by cells in the other published cohorts and in the GBO scRNA-seq libraries. We extracted a raw-counts matrix including the intersection between genes detected in the cohort and the top 4,000 variable genes in the MGB cohort. This matrix was then subjected to the cNMF prepare script for normalization. The –numgenes parameter was set to the number of genes in the matrix. We used sklearn.decomposition.non_negative_factorization in which X is the filtered normalized expression matrix and H is the filtered gene-spectra consensus matrix. The following parameters were used: n_components= 18, init = ‘random’, update_H=False, solver=‘cd’, beta_loss=‘frobenius’, tol=0.0001, max_iter=1000, alpha=0.0, alpha_W = 0.0, alpha_H = ‘same’, l1_ratio=0.0, regularization=None, random_state=None, verbose=0, shuffle=False. The code is available at https://github.com/BernsteinLab/Myeloid-Glioma.
Finally, each cell was annotated as a cell type using the final ‘usage matrix output of cNMF or the calculated usage matrices, as discussed above. The usage scores were normalized to 100% for each cell. For each cell, the usage scores for all programs in each category were summed to create a usage score for the cell-type category. For example, the usage scores for four myeloid programs were summed to create the myeloid usage per cell. Cells were then annotated as one of the cell types using the top-scoring usage for cell-type category.
Of note, cycling cells were considered separately. We used inferCNV to annotate cycling cells as malignant or non-,alignant. Non-malignant cells were then further annotated by the next-highest cell type. These secondary annotations were used when separating cell types for further cell-type-specific analysis.
CNA inference from single-cell data
We selected a group of reference cells not annotated as any malignant program from various tumours (a mix of myeloid cells, T cells, oligos and vasculature cells). We extracted and merged the raw counts of these reference cells into a single matrix. The reference cells used are given in https://github.com/BernsteinLab/Myeloid-Glioma. We then used the inferCNV package (inferCNV v.1.3.3 of the Trinity CTAT Project; https://github.com/broadinstitute/inferCNV). We performed the analysis for each tumour separately. In the annotation file, we included the reference cells and annotated the cells of each tumour, as discussed above. We concatenated the raw matrix of each tumour with the reference cells’ raw matrix. We constructed the gene-order file required for inferCNV using the gtf_to_position_file.py script of the inferCNV package. We included the following extra arguments: –denoise –HMM –cluster_by_groups –cutoff 0.1. We also ensured that the –ref_group_names match the names given to the reference cells in the annotation files. The selection of the reference cells was performed separately for each cohort.
Doublet detection
Doublets were determined using integration of cNMF and inferCNV data. Cells were considered doublets by cNMF if they expressed a second program above a specific threshold (Supplementary Table 6). Cell-type-specific thresholds were selected by subsetting by cell type, then plotting the usage of each potential second program. From this plot, we found the value that separated the background usage of a second program from doublets. Cells were also considered doublets if their cNMF annotation was not compatible with the inferCNV profile. Of note, the cycling programs were not considered in doublet analysis. A more detailed description of annotation and doublet-detection methods can be found at https://github.com/BernsteinLab/Myeloid-Glioma/blob/main/Processing%20of%20scRNA-Seq%20Files%20(Related%20to%20Figure%201)/15-%20Annotation%20and% 20Doublet%20Detection.pdf.
Integrated definition of malignant cells
If a tumour had CNVs that were detectable using inferCNV, cells from that tumour had to meet the following criteria: non-doublet, positive for CNV and not annotated as a non-malignant cell type by the cNMF program. For those tumours in which CNVs could not be readily detected by inferCNV, we relied on annotations based on cNMF.
Gene-program identifications
For more granular analysis of cell programs for a specific cell type (myeloid cells, T cells or malignant cells), we took cells in each specific category and removed doublets on the basis of the method described in the ‘Doublet detection’ section above. We then input those cells identified as singlets into another cNMF analysis for each category.
Myeloid cells
We used the MGB, Jackson Laboratories and Methodist cohorts for identifying the cNMF programs in myeloid cells in gliomas. The cNMF was carried out in two rounds for each cohort. The first round was used to identify cells using programs that are not myeloid (that is, have a different cell-type identity) or programs used by fewer than 100 myeloid cells. We removed such cells for subsequent analyses. The second round was used to determine the myeloid programs.
In the first round, raw counts of all cells annotated as myeloid and singlets (non-doublets) from each cohort were used to create a Seurat object independently. We then normalized the Seurat object using NormalizeData() and identified the top 2,000 variable genes with mean expression greater than 0.001 in expressing cells in each cohort using the FindVariableFeatures(). Subsequently, we output the three matrices. These matrices were subjected separately to cNMF with the following parameters in the prepare script: –n-iter 500 –total-workers 1 –seed 14 –numgenes 2000. Then we performed factorization and generated the K-plots using the factorize, combine and k_selection_plot scripts of cNMF. We then chose the following Ks: MGB – 22, Houston Methodist – 23, Jackson’s laboratories – 14. We then performed the consensus script with the above Ks and a local-density threshold of 0.02.
In the second round, we removed cells from each cohort as discussed above and created a merged Seurat object from the three cleaned matrices using the Seurat merge() function. Then we normalized the merged Seurat object and detected variable genes using NormalizeData() and FindVariableFeatures(). We then filtered out the genes with a mean expression value of less than 0.01 in expressing cells and standardized variance of less than 1. We then filtered the cleaned myeloid matrix of each cohort to include the variable genes that met the criteria mentioned above. Similar to round 1, these matrices were subjected to cNMF individually with the following parameters in the prepare script: –n-iter 500 –total-workers 1 –seed 14 –numgenes 2276. Then we also ran the factorization and generated the K-plots using the factorize, combine and k_selection_plot scripts of cNMF. We then chose the following Ks in the second round: MGB – 18 (we filtered out programs that are not myeloid), Houston Methodist – 19, Jackson Laboratories – 18. Finally, we performed the consensus script with the above-mentioned Ks and a local-density threshold of 0.02.
To find the consensus programs, we performed a cosine similarity of the gene-spectra ‘dt’ output of each discovery cohort from round 2 of cNMF. We applied a hierarchical clustering algorithm called Ward’s method to the similarity matrix to visualize the relationships between programs in a heatmap. To derive the spectra scores for the consensus programs, we averaged the spectra dt scores of the programs with a cosine similarity score of 0.5 or higher. This resulted in 14 consensus myeloid programs across the 3 cohorts, which we annotated as discussed above.
Malignant and T cells
Malignant cells and T cells were obtained from the MGB data in separate cNMF runs, similar to the two-step cNMF used for myeloid cells. We selected a K-value of 7 for the malignant cells based on the silhouette plot’s stability, consistent with previously published glioblastoma signatures represented in our five chosen programs61. For the T cells, we found the optimal program count to be 4. We calculated the usage of these programs in the other cohorts in a way similar to the all-cell-type NMF mentioned above.
Processing and NMF for PBMC scRNA-seq libraries
The PBMC libraries were processed for cNMF in a similar way to the primary tumour libraries. We merged the expression matrix of all the PBMC libraries using the Seurat merge function. The Seurat object was then normalized using NormalizeData() and ScaleData(). We then used FindVariableFeatures() to calculate the variance score for every gene. We selected the top 3,000 variable genes after removing genes of less than 0.001 mean expression (in expressing cells) and then subsetted the gene-expression matrix to include only the variable genes. As described above, cNMF was performed with –numgenes 3000 and a value K = 18 for the consensus script of cNMF, annotation was done using gProfiler, and non-doublet cells were identified. We isolated myeloid cells, identified the top 2,000 most-variable genes, and performed two rounds of cNMF (K = 16).
Calculation of the usage of myeloid programs
We extracted a raw counts matrix including the intersection between genes detected in the GBO matrix or other tumour types of matrices and the genes found in the myeloid-cell programs gene-spectra file. This matrix was then subjected to the cNMF prepare script for normalization. The –numgenes parameter was set to the number of genes in the matrix. We used sklearn.decomposition.non_negative_factorization in which X is the filtered normalized expression matrix and H is the filtered gene-spectra consensus matrix. The following parameters were used: n_components= 14, init=‘random’, update_H=False, solver=‘cd’, beta_loss=‘frobenius’, tol=0.0001, max_iter=1000, alpha=0.0, alpha_W=0.0, alpha_H=‘same’, l1_ratio=0.0, regularization=None, random_state=None, verbose=0, shuffle=False. The code is available at https://github.com/BernsteinLab/Myeloid-Glioma.
The usage scores were normalized to 100% for each cell.
Comparison of gene programs
To assess the similarity of 2 given gene programs, we took the top 100 genes in those programs and compared their make-up using a Jaccard index. P-values were measured by assessing the probability of observed gene matches being obtained by random chance using a binomial test in which k is the number of matches, n is the size of the gene set, and P is probability of randomly drawing matches from all genes scored in the program.
Comparative UMAP and clustering of myeloid cells
We extracted the raw counts of all MGB cells annotated as myeloid and singlets (non-doublets) from each tumour. Then normalization was performed for each tumour separately using NormalizeData() with default settings, followed by FindVariableGenes() with the following settings (selection.method = vst, nfeatures = 2,000). We then ran FindIntegrationAnchors() k.filter set at 30 (to ensure that tumours with few myeloid cells were included. We then used the anchors identified as input to batch correct the objects using IntegrateData(), setting features.to.integrate as the intersection of genes detected in all tumours in and dims to 1:30.
The batch-corrected Seurat object was then subjected to ScaleData(), RunPCA() and ElbowPlot() with default parameters to identify the number of dimensions to use for Louvain clustering and UMAP generation. We generated the UMAP using RunUMAP() with dims set to 1:8 and reductions set to pca. We performed the clustering using FindNeighbors() with dims set to 1:8, followed by FindClusters() with a 0.3 resolution. UMAPs were generated using the DimPlot() function.
Generation of heatmap for gene-expression programs
To generate the gene-expression heatmap of the NMF programs, we assigned the myeloid cells to one of the following categories:
Microglia
This had a minimum 10% usage of the microglia program and other identity programs were all less than the usage value of the microglia program (macrophages must be below 10%).
Microglia-like
This had a minimum 10% usage of microglia and 10% usage of the monocytes or macrophages program. Other identity programs should be below the usage value of these two programs (otherwise it was assigned as a microglia).
Macrophages
This had a minimum 10% usage of the macrophage program and other identity programs were all below the usage value of the macrophage program (monocytes below 10%).
Mono_macro
This had a minimum 10% usage of macrophages and 10% usage of the monocytes program. Other identity programs were below the usage value of these two programs.
Monocytes
This had a minimum 10% usage of the macrophage program and other identity programs were below the usage value of the monocytes program.
cDC
This had a minimum 10% usage of the cDC program and other identity programs were all below the usage value of the cDCs program.
Neutrophils
This had a minimum 10% usage of the neutrophils program and other identity programs were all below the usage value of the neutrophils program.
Activity dominated
All identity programs were below 10% usage.
For selecting the genes included in the heatmaps, we identified the top 100 genes in the averaged gene-spectra output of the myeloid cNMF programs for each program. We counted the number of myeloid cells expressing the top 100 genes in each program. We included the top 20 genes with the highest number of myeloid cells expressing them in each program.
We used the ComplexHeatmap library in R63 to generate the heatmaps. We z-score scaled the log1p normalized gene-expression values across all the myeloid cells (regardless of the categorization). We then set an upper limit of 2 and a lower limit of −1. We generated a heatmap for each category separately. We turned off row clustering (genes) by setting cluster_rows = FALSE in the heatmap function, and we allowed default column clustering in each category (cells).
Generation of quadrant plots
We plotted 183,062 myeloid cells from 85 tumours on the basis of their expression of the four immunomodulatory programs, orienting largely exclusive inflammatory and immunosuppressive programs oppositely on the same axes. The x axis of the quadrant plots (bottom left to top right) was calculated by subtracting the usage of the complement immunosuppressive program from the systemic pro-inflammatory program. The y axis (top left to bottom right) was calculated by subtracting the scavenger immunosuppressive program usage from the microglial inflammatory program for each myeloid cell. The resulting coordinates were transformed using a 45° rotation matrix. For the quadrant plot with scatterpies as dots, we used the scatterpie library (https://github.com/GuangchuangYu/scatterpie). The ‘others’ category for the pie charts was calculated by summing the usages of all programs excluding the four immunomodulatory programs.
Determining cut-off usage values for myeloid cells
To determine the cut-off threshold for myeloid identity programs in primary tumours, we created an elbow plot to analyse the usage values for each myeloid-identity cNMF program. Based on our findings, we identified the minimum inflection point for an identity program at around 9% and set the cut-off level at 10% usage. Similarly, we observed that the minimum inflection point is approximately 18% for activity programs, leading us to select 20% usage as the cut-off level. A single myeloid cell could be classified using multiple programs. For example, a myeloid cell could be considered to be microglia using the scavenger immunosuppressive program if it had at least 10% usage of the microglia program and 20% usage of the scavenger immunosuppressive program. These cut-offs were used for Fig. 1d and Extended Data Figs. 1e and 8b. For Extended Data Fig. 2d, we calculated the observed/expected ratios of the co-occurrence of a myeloid identity program and a myeloid activity program and used a hypergeometric test to assess significance.
For GBOs, as part of optimizing the model, we determined the inflection points of the activity programs in the myeloid cells. We observed that the inflection points were approximately 27% for activity programs, leading us to select 30% usage as the cut-off level in GBO experiments. These cut-offs were used for all GBO scRNA-seq analyses, as shown in Fig. 5d and Extended Data Fig. 12a.
Definite annotation for myeloid cells and T cells
Specific analyses (MAESTER, Extended Data Fig. 5; and heatmap generation, Fig. 1c) required definite annotation of a myeloid cell instead of only considering whether the myeloid cell uses a particular program or not. The cut-offs for such analyses are mentioned in their respective sections.
T cell program usages were more distinct. We therefore simply defined them by the program of their top usage.
Creating discretized matrix and identifying marker genes
To enable the discovery of specific markers and the spatial cellular demultiplexing described below, we created a discretized matrix of MGB cells with the strongest expression of each tumour-cell program, including myeloid, T cell and cancer programs, thereby excluding intermediate cells. For the outputs of myeloid, malignant and T cell cNMFs, cells with a minimum 2.5-fold higher usage of a particular program over the second-most-used program were annotated with that program as a discrete cell. For oligodendrocytes and vasculature, the usages from all cell types cNMF outputs were used to annotate oligo or vasculature discrete cells. Doublets, cycling programs and cycling cells were excluded from the analysis.
Furthermore, we downloaded scRNA-seq for normal brain tissue from individuals 25 and 40 years old64. These libraries were processed using the above-mentioned Seurat processing; we normalized the cells and used 1:14 as dims for generating UMAPs and identifying clusters. We performed FindMarkers and extracted neurons and astrocytes from the published matrix on the basis of differentially expressed genes. We then merged these cells with the discrete cells matrix. We generated a UMAP as described above.
To identify markers for the immunomodulatory programs, we extracted discrete cells annotated as scavenger immunosuppressive, complement immunosuppressive, systemic inflammatory or microglial inflammatory using the Seurat subset() function (discrete myeloid immunomodulatory object). A similar processing pipeline was performed with the option dims set to 1:16 in FindNeighbors() and FindClusters(). The UMAP coordinates for these cells were obtained using Embeddings() with the option reduction set to umap. Then the normalized matrix of these cells was extracted using the function GetAssayData() with the option slot set to data. These files were used as input for COMET65 to identify the significant markers that distinguish each immunomodulatory program. We also used the Seurat function DotPlot() to generate the expression dot plots shown in Extended Data Fig. 10c.
Regulon analysis
We used the normalized discrete myeloid immunomodulatory expression matrix as an input for SCENIC41. We also downloaded the feather files hg38_10kbp_up_10kbp_down_full_tx_v10_clust.genes_vs_motifs.rankings.feather and hg38_500bp_up_100bp_down_full_tx_v10_clust.genes_vs_motifs.rankings.feather from https://resources.aertslab.org/cistarget/databases/homo_sapiens/hg38/refseq_r80/mc_v10_clust/gene_based and used these as input for SCENIC. Genes in the matrix were filtered on the basis of the default settings, and we ran the four steps runSCENIC_1_coexNetwork2modules, runSCENIC_2_createRegulons, runSCENIC_3_scoreCells(scenicOptions, exprMat_filtered) and runSCENIC_4_aucell_binarize. To calculate the regulon specificity score for each regulon in each immunomodulatory annotation, we used the function calculate_rrs() of scFunction (https://github.com/FloWuenne/scFunctions/). The full codes can be found at https://github.com/BernsteinLab/Myeloid-Glioma.
MAESTER analysis for determining myeloid-cell origin
To determine the origins of the various myeloid-cell identities, we processed the MAESTER libraries in three stages: preprocessing, identifying variants of interest, and measuring the enrichment of the identified variants in the various myeloid identities.
Stage 1: preprocessing. The preprocessing was done as previously described30. In brief, we trimmed high-quality reads, aligned them using STAR (hg38) and processed them using MAEGATK with the default settings except for – -min-reads, which was set to 3 to ensure a minimum of three reads to cover the base in the cell30.
Stage 2: low-resolution pseudobulking to identify variants of interests. To determine the origin of the myeloid-cell identities, we identified variants specific to myeloid cells in the tumour microenvironment but not present in the PBMC. We also identified variants present in the myeloid cells in the PBMC but absent in the tumour microenvironment. We pseudobulked the primary tumour libraries to the following categories according to their RNA-seq annotation. For the primary tumour libraries Malignant, Tumor-Associated Myeloid (TAMs), Stromal, Oligo and Tcells, and for PBMC libraries, we considered only Neutrophils and Monocytes (Myeloid_PBMCs). This stage involved multiple steps.
Step 1: we extracted the number of UMIs supporting each possible variant at every possible location from the MAEGATK output using the following script in R:
computeAFMutMatrix <- function(SE){
ref_allele <- as.character(rowRanges(SE)$refAllele)
getMutMatrix <- function(letter){
mat <- (assays(SE)[[paste0(letter, “_counts_fw”)]] + assays(SE)[[paste0(letter, “_counts_rev”)]])
rownames(mat) <- paste0(as.character(1:dim(mat)[1]), “_”, toupper(ref_allele), “>”, letter)
return(mat[toupper(ref_allele)!= letter,])
}
rbind(getMutMatrix(“A”), getMutMatrix(“C”), getMutMatrix(“G”), getMutMatrix(“T”))
}
The computeAFMutMatrix generated a matrix in which the rows are every possible variant in the mitochondrial genome, and the columns are barcodes (cells). The values represent the UMIs supporting each variant in the given barcode.
Step 2: we extracted the coverage of the Maester libraries at each base of the MT genome in every cell from the output of MAEGATK, which was stored in assays(maegatk.rse)[[coverage]], whereby maegatk.rse is the R object output of MAEGATK.
Step 3: we annotated the cells in each matrix (obtained from step 1 and step 2) based on the scRNA-seq library (as mentioned in the ‘Classification of cell types’ section). We created a data frame in which one column has the barcodes and the other has the annotation.
Step 4: we pseudobulked the UMI count matrix (obtained from step 1) using R with the following steps: we subsetted the matrix into each annotation using tibble; we used the sum() function to sum all the rows in each matrix, creating a pseudobulked number for each annotation; and we merged all the summed values in each matrix for each variant possibility into a pseudobulked matrix in which each column represents an annotation.
Step 5: we pseudobulked the coverage matrix (obtained from step 2) similarly to step 4.
Step 6: we calculated the pseudobulked variant allele frequencies (VAFs) using R. We added a pseudo-count 0.000001 to each value in the pseudobulked coverage matrix. We divided each value in the pseudobulked counts matrix by its coverage in the pseudobulked coverage matrix to obtain pseudobulked VAFs for each category.
Step 7: to ensure the specificity of each variant’s detection with 99% certainty, we used a binomial model to establish a minimum VAF threshold dependent on coverage.
Step 8: to consider the variant to be specific to the myeloid cells in the tumour microenvironment, the variant had to meet the following criteria: the minimum VAF requirement for TAMs for the coverages in TAMs and Myeloid_PBMC categories; VAF = 0 in the myeloid PBMC category; and VAF > the minimum required in the TAM category.
Step 9: to identify variants specific to the myeloid cells in PBMCs, the variant had to meet the following criteria: first, it had to achieve the minimum VAF requirement for Myeloid_PBMC for the coverages in the Malignant, TAMs and Myeloid_PBMC categories; second, VAF = 0 in the Malignant category (if the tumour library was enriched for malignant cells, this criteria could be replaced with VAF in Myeloid_PBMC, which is 20 times bigger than Malignant); third, VAF > 0 in the TAM category; and fourth, VAF > the minimum required in the Myeloid_PBMC category. The mutations used for subsequent analyses can be found in Supplementary Table 7.
Stage 3: determining the enrichment of variants of interest in various identities of myeloid cells.
We hypothesized that myeloid cells enriched for tumour microenvironment-specific variants were tissue residents, whereas those enriched for PBMC-specific variants were derived from monocytes. Performing the enrichment analysis had several stages. We began by annotating the TAMs with the myeloid identity cNMF programs. We kept cells that met the following criteria to ensure the reliability of the identity and the results:
for Microglia annotation: first, the cell has a minimum of 15% usage of microglia program; second, the microglia program must be at least twice as high as any other identity program; and third, the macrophage program is at least twice as low as any other identity program.
for Microglia_Like annotation: first, the cell has a minimum of 10% usage of the microglia program; second, the cell has a minimum of 10% usage of the macrophage program; third, the microglia program must be at least twice as high as any other identity program (excluding macrophages and monocytes); and fourth, the macrophage program must be at least twice as high as any other identity program (excluding microglia and monocytes).
for Mono_Macro annotation: first, the cell has a minimum of 10% usage of the monocytes program; second, the cell has a minimum of 10% usage of the macrophage program; third, the monocyte program must be at least twice as high as any other identity program (excluding macrophages); and fourth, the macrophage program must be at least twice as high as any other identity program (excluding monocytes).
for Macrophages annotation: first, the cell has a minimum of 15% usage of the macrophages program; second, the macrophage program must be at least twice as high as any other identity program; third, the monocytes program must be at least twice as high as any other identity program; and fourth, the microglia program must be at least twice as high as any other identity program.
For Monocytes annotation: first, the cell has a minimum of 15% usage of the monocytes program; second, the monocyte program must be at least twice as high as any other identity program; and third, the macrophage program must be at least twice as high as any other identity program.
For Neutrophils annotation: first, the cell has a minimum of 15% usage of the neutrophils program; and second, the neutrophils program must be at least twice as high as any other identity program.
For cDC annotation: first, the cell has a minimum of 15% usage of the cDC program; and second, the cDC program must be at least twice as high as any other identity program.
We subsetted the single cells MAESTER UMI matrix obtained in stage 2, step 1 to smaller matrices, with each matrix composed of cells annotated with one of the above identities (stage 3, step 1).
We created a matrix in which the rows are the variants of interest identified in stage 3, and each column represents an identity. The values in this matrix denote the number of cells in each identity in which the variant was detected. We used annotation_matrix[,apply(annotation_matrix,2,function(x) sum(x > 0))] in R for each subsetted matrix. This script keeps columns that have UMI > 0. Then we identified the number of cells for which UMI > 0 for each annotation by using ncol(as.matrix(annotation_matrix), whereby annotation_matrix represents each subsetted matrix.
We then removed any variant with a zero value in every pseudobulked category.
We removed any identity (column) that had less than ten total values to ensure proper enrichment results.
We used GSVA66 by categorizing the rows of the matrix as PBMC specific or tumour microenvironment specific. Because the values are integers (the number of cells having UMIs supporting the variants), we set the option kcdf to Poisson in the gsva script.
We then subtracted the GSVA enrichment for tumour microenvironment-specific variants from GSVA enrichment for PBMC-specific variants for each identity. Positive values indicate a PBMC origin, whereas negative values indicate a tissue-resident origin.
We calculated the prevalence of each identity in each tumour. We plotted the dot plots with the position of the dots representing the subtracted GSVA enrichment values, and the size of the dot representing the prevalence using ggplot2.
We calculated the average percentage usages of the four immunomodulatory programs in each identity, labelled the remaining percentage as others in each tumour, and plotted them as stacked bar plots using ggplot2.
Sample level association analysis
To measure inter-state association, we determined the average usage of the myeloid, T cell and cancer programs in their respective cells in each sample. We used the Spearman correlation to understand how a rise in one myeloid state corresponded to changes in cancer or T cell states, enabling us to examine how variations in one state could influence the behaviour of others in the same sample.
In our state-clinical/molecular associations analysis, we applied the sample state averages derived above. These were compared across various clinical and molecular categories, such as tumour grade, IDH-mutation status and steroid usage, using the Wilcoxon rank-sum test.
To account for the confounding effect of sample hypoxia on the influence of dexamethasone on the complement immunosuppressive myeloid program, we limited this analysis to samples for which the average MES2 program usage in cancer cells was below 20%.
We used a linear least-square regression to investigate changes in average myeloid program usage across samples relative to the daily dose of Dex.
To assess the myeloid and cancer program associations with Treg cells, we classified each sample in the top 50% and bottom 50% expressors of Treg cells and cancer/myeloid program usages and used Fisher’s exact test to measure this association. P-values were adjusted using the FDR.
Cell-cycle analysis
We evaluated the relationship between the cell cycle and different myeloid cell states in each sample independently. Cells were defined as cycling if the combined usage of the G1S and G2M programs exceeded 20%, and as belonging to a specific state if its usage surpassed 20%. We used Fisher’s exact test to measure this association and obtained an odds ratio. For each program, we assessed the probability that its sample distribution was higher than 1 using a one-sample t-test on the log-transformed values, so they were approximately normally distributed67.
Deconvolution of TCGA, GLASS and G-SAM
The analysis pipeline consisted of three steps to deconvolve previously published glioma bulk expression datasets. Steps 2 and 3 of the pipeline were performed for each dataset separately.
Step 1: creating gene sets for each program. The top 50 genes were obtained for each myeloid program by ranking them in the merged gene-spectra output. For T cells and malignant cells, we ranked the gene spectra from their respective cNMF outputs to obtain the top 50 genes for each program. For the other cell types, we used the gene-spectra output of the cNMF of all cells in the tumour microenvironment. We ranked and obtained the top 50 genes for the pericytes, endothelial and oligo programs. Then we selected genes that appeared only in the list of a single program.
Step 2: calculating module scores using Seurat. Raw gene counts for the glioma datasets were obtained. The matrices were then CPM-normalized using EdgeR’s DGEList(), calcNormFactors() and cpm() functions68. The CPM-normalized matrix was then log-transformed using the log1p() function of R. Afterwards, Seurat objects were created using the CreateSeuratObject() function. The Seurat objects were scaled using ScaleData(). Finally, module scores were calculated using Seurat’s AddModuleScore() function. The above-mentioned gene sets were used as input in the features option of AddModuleScore().
Step 3: normalizing the module score. To correct for the purity differences in the published bulk glioma mRNA-expression datasets, we imputed the percentages of malignant, oligo, vasculature, myeloid, T cells and other immune cells in the bulk expression matrices using CIBERSORTx69. As input, we used the discretized matrix described above, excluding neurons and astrocytes. The matrix was used as the single_cell_reference in the Fractions module of CIBERSORTx. The published bulk gene expression matrices used as ‘mixture’ input were also CPM-normalized without log transformation. The module scores obtained above were then divided by the imputed value of their respective cell type in the CIBERSORTx results outputs.
These normalized module scores were then used to correlate with sample clinical data, including IDH-mutation status and molecular grade. For correlation with clinical data, the normalized module scores were log10 transformed. We removed libraries with a CIBERSORTx value of less than 0.1 for myeloid cells. To correlate the module scores with the expression levels of the identified ligands in the lower-grade glioma and glioblastoma TCGA cohorts, we performed a Pearson correlation between the log-transformed normalized module score and the log-transformed normalized expression. We used the Wilcoxon rank sum test to assess the statistical significance, adjusting the P-values using FDR.
Spatial transcriptomic analysis
We used Visium spatial transcriptomics to analyse 23 samples32. Our methodology encompassed two distinct approaches: unbiased niche discovery using cNMF; and cell-state demultiplexing using RCTD33 and the single-cell RNA-seq programs.
In the unbiased niche discovery, we used all samples, both healthy and cancer, for cNMF analysis. We selected the 1,500 top spatially variable genes based on Moran’s I and ran the cNMF algorithm from k = 2 to k = 25. The optimal k value (k = 7) was determined as the highest one with a silhouette score greater than 0.9. We defined the identity of niche programs through the analysis of gene expression patterns and excluded the program whose top genes were mitochondrial and ribosomal. To ensure uniformity, the usage scores were normalized to 1 in each spot.
For cell-state demultiplexing, we used the discretized matrix described above. Using this single-cell reference, RCTD was run individually for each spatial sample. Again, the scores were normalized to sum to 1 in each spot.
To assess the link between niche and cell state, we used the Spearman correlation to evaluate the relationship between normalized RCTD scores and cNMF usage for individual patient samples. For Fig. 2, a cell type was classified as part of a niche if the correlation had statistical significance (FDR < 0.01) in more than half of the samples.
In our analysis of cell state and niche–niche spatial relationships, we designed a spatial regression model to quantify cell-state proximity, analysing one sample at a time. For any pair of cell or niche states across all spots in a sample, we regressed the presence of one state in a central spot against the average expression of the other state in surrounding spots. In this model, we binarized the central spot’s state presence using a state-specific threshold determined by a knee plot (KneeLocator70) of cell-state expression across spots in all samples. The resulting regression coefficient indicates the association strength between the states, with the P-value demonstrating the statistical significance of this association. The constant represents the background signal of the non-central state. The network graph in Extended Data Fig. 6f was plotted using the Python package NetworkX (v.2.0). The size of each edge represents the ratio of the regression coefficient to the constant, providing a measure of the state-association strength relative to the background signal. Edges were statistically significant (FDR < 0.01) in more than 40% of the samples with an association strength greater than 0.1.
Correlation of immunomodulatory programs with responders and non-responders to immunotherapy
We downloaded the published Seurat object of scRNA-seq libraries of patients with glioma responding or not responding to immunotherapy38. Based on the published annotations, we extracted the myeloid cells from the Seurat object. We calculated the usage of the myeloid cNMF programs in each myeloid cell, as shown above. We averaged the usage of each program in the myeloid cells per patient and calculated the difference between the average usage in responding versus non-responding patients, using FDR-corrected Wilcoxon rank-sum tests to assess the significance.
We calculated the module scores using gene sets from refs. 7,15,19 in all myeloid cells from ref. 38. Similarly, we averaged the module scores of each gene set for each patient and calculated the difference between responding versus non-responding patients, using FDR-corrected Wilcoxon rank sum tests to assess the significance.
Survival analysis
We merged the survival information (survival time) and events from the cohorts of G-SAM and GLASS. We included only IDH-WT glioblastomas. We used the normalized module score values (without log transformation) for the cNMF programs. We removed duplicate patient entries corresponding to recurrences by keeping the values from the primary tumour only. We removed any library with a CIBERSORTx value of 0 for the myeloid lineage. We then took the samples in the top 33% and considered this group of samples as the high in each program. For the low group in each program, we selected samples in the bottom 33% of libraries.
The library ggsurvfit (https://github.com/pharmaverse/ggsurvfit) was then used to generate the Kaplan–Meier survival curve. A Cox proportional hazard model (https://github.com/therneau/survival) was used to determine differences in survival probabilities.
We performed the survival analysis in a similar way using gene sets from refs. 7,15,19.
Single-cell ATAC-seq analysis
We downloaded 20 snATAC-seq libraries from ref. 40 and the fragments file for each library. Each library was then processed using Signac71 v.1.12.0 to remove low-quality cells. In brief, a fragment object for each fragment file was created using the CreateFragmentObject() function of Signac. Then, peaks were called using the CallPeaks() function. We removed peaks mapping to contigs by using keepStandardChromosomes() and removed blacklisted regions by using the subsetByOverlaps() function with the options ranges = blacklist_hg38 and invert = TRUE.
We created a counts matrix for each library using FeatureMatrix(), then created a chromatin assay by inputting the created counts matrix and fragments file of the respective library using the CreateChromatinAssay() function of Signac. We filtered cells that did not include reads in more than 200 features and removed peaks that did not include reads from more than 10 cells. Finally, the Seurat object for each snATAC-seq library was created using the CreateSeuratObject() function.
The cells of each library were subject to QC analyses using the NucleosomeSignal(), TSSEnrichment(), FractionCountsInRegion() and FRiP() functions to filter out cells with high nucleosomal noise, those high in reads mapped to blacklisted regions, low enrichment near TSS and/or low in the number of reads contributing to peaks. Thresholds selected for each library can be found at https://github.com/BernsteinLab/Myeloid-Glioma. The Seurat object and peaks for each library were saved after filtering out low-quality cells.
Next, we combined all the Seurat objects into one as follows. First, we merged all the peaks using the Reduce() function and filtered out peaks less than 20 base pairs ( bp) or more than 10,000 bp in length. Then we created a new fragment object for each library by inputting the original fragments file into CreateFragmentObject() but used only high-quality cells. We then created a counts matrix for each library by inputting the newly created fragment objects and combined peaks using the FeatureMatrix() function. Then we created new chromatin assays for each library by inputting the newly created counts matrices and fragment objects using the CreateChromatinAssay() function. Next, we created a new Seurat object for each library using the newly created chromatin assays. Finally, we combined the new Seurat objects using the merge function.
The combined Seurat object was then subjected to clustering analysis and UMAP generation using the RunTFIDF(), FindTopFeatures(), RunSVD(), RunUMAP(), FindNeighbors() and FindClusters() functions. We used the lsi reduction and removed PC1 and PC3 because they were associated with library depth, not biology. The function DepthCor() was used to determine the association of each PC with depth.
The gene activities were calculated using the GeneActivity() function of Signac. Then we created a gene activity matrix using CreateAssayObject(). The values of gene activities were normalized using the NormalizeData() function with the options normalization.method set to RC and scale.factor set to median(Object$nCount_RNA).
The gene activity matrix was then subject to calculation of the cell-types cNMF programs, as described above for the scRNA-seq matrices, to annotate the cells in the matrix. Using these calculations, the non-doublet myeloid cells were identified in the combined Seurat object as mentioned above.
The Seurat object was filtered to include only myeloid cells by using the subset() function. After that, a new set of peaks was called, resulting in the creation of a new feature matrix and a new chromatin assay. Finally, a gene activity matrix was generated for the combined myeloid-cells object, as discussed previously. The usage of the myeloid programs was determined in the myeloid cells to extract the discrete myeloid cells, as mentioned above, for the scRNA-seq libraries. We extracted discrete myeloid cells for monocyte and microglia identities and for systemic, scavenger, complement and microglial inflammatory for immunomodulatory activities. For systemic inflammatory myeloid cells, we selected the top 400 discrete myeloid cells.
To determine differentially accessible peaks in the immunomodulatory programs, we pseudobulked the discrete myeloid populations using the AggregateExpression() function with the following options: return.seurat = TRUE, group.by = Myeloid_Discrete_Status4, normalization.method = LogNormalize, scale.factor = 10,000. Then we extracted the normalized counts matrix using the LayerData() function on the pseudobulked Seurat object with the layer option set to data. We then converted the log1p values to exponential values and considered a peak to be specific to a particular discrete annotation if it had a count at least 2.5 times greater than the average counts of the other annotations. We generated normalized bigwigs for each annotation using an in-house code, which can be found at https://github.com/BernsteinLab/Myeloid-Glioma. To generate a reference matrix of coverage for the regions spanning 1,000 bps upstream and downstream of the centres of the specific peaks, we utilized the deeptools computeMatrix function. We inputted the normalized bigwigs and the specific peak files while enabling the reference-point and −skipZeros options, and setting –referencePoint to centre and −b and −a to 1,000. Finally, we plotted the heatmap using the deeptools plotHeatmap function72.
We performed motif analysis on each specific peak set using monaLISA73 v.1.11.4 to identify significantly enriched motifs. First, we combined the specific peaks and resized them to the median width of the combined peaks with a centre fixation. Subsequently, we divided the combined peak files into bins based on the annotation of the peak. We used the getSeq() function to obtain the sequences of the peaks. We used the PWM core JASPAR2024 (ref. 74) as features and determined the motifs using the calcBinnedMotifEnrR() function of monaLISA. To create a large background for calculating enrichments, we set the background to hg38 genome and the genome.oversample = 500. Finally, the motifs heatmap was generated using the plotMotifHeatmaps() function.
Ligand–TF analysis
To understand which ligands are involved in driving specific programs and regulatory circuitry, we used the NicheNet42 database (https://zenodo.org/record/7074291/files/gr_network_human_21122021.rds) of file names gr_Network_Nichenet_Database.txt or Signaling_Network_Nichenet_Database) that connects ligands to the TFs they can activate through known ligand–receptor interactions.
We first identified a list of potentially active TFs for each immunomodulatory program through motif enrichment analysis of specific ATAC-seq loci (Fig. 4a,b) and SCENIC analysis (Fig. 4c). Then we used the NicheNet database to identify all ligands that were connected to each TF. We kept only ligands that were expressed in cells in the tumours (any cell type in the tumour microenvironment) at more than 0.3.
Bulk ATAC-seq analysis
The Nextera transposase adaptors were removed from the fastq files by trimming them with the –phred33 –paired –fastqc options using trim_galore75 (https://github.com/FelixKrueger/TrimGalore). The STAR aligner60 was then used to map the fastqs with the following settings: -outSAMtype BAM SortedByCoordinate –outFilterMultimapNmax 1000 –outFilterScoreMinOverLread 0.25 –alignIntronMax 1 –alignEndsType EndToEnd. We then used the PICARD MarkDuplicates module (https://broadinstitute.github.io/picard/) with the option REMOVE_DUPLICATES set to TRUE. Afterwards, samtools76 was used to remove reads mapped to chrM or contigs. The processed bam files were then used to generate bigwigs using the bamCoverage function of deeptools with the RPGC normalization method and an effective genome size of 2,913,022,398. To identify accessible regions, we converted the processed bam files to bed format using bedtools77 and used awk to add four bases to the start site if the read was on the positive strand and remove five bases from the end site if the read was on the negative strand to correct for tn5 biases. MACS2 v.2.2.9.1 was used to call peaks using the following settings: -g hs -f BED -q 0.01 –nomodel –shift -75 –extsize 150 –keep-dup all. To identify differential accessible sites, we created tag directories using the processed mapped files as input for the makeTagDirectory function of HOMER78. The accessible sites for DMSO and p300i libraries were merged using the mergePeaks function of HOMER, and differential accessible sites were determined using the getDifferentialPeaks function of HOMER and setting -F to 2 to keep regions with two-fold higher normalized tag counts in one of the libraries. Deeptools heatmaps were generated as mentioned above, and significant motifs enriched in the differential accessible sites were identified using monaLISA v.1.11.4, similar to the way mentioned above, except for setting genome.oversample to 50, given the number of differentially accessible sites.
GR ChIP–seq analysis
The binding sites of GR were obtained from ref. 43. The binding sites were annotated using the findPeaks module of HOMER. The maximum allowable distance to a gene was 100,000 bp.
Temporal correlation analysis
We obtained gene sets for genes correlated with recent infiltration and remote infiltration into the tumour in a mouse model of glioblastoma50. We used gprofiler (https://biit.cs.ut.ee/gprofiler/gost) to determine the enrichment of the myeloid cNMF programs in these gene sets by submitting the top 100 genes for each cNMF program as a query against each gene set as a background.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.