Preparation of expression vectors
Plasmids are listed in Supplementary Table 1, oligo sequences in Supplementary Table 2 and gBlocks (Integrated DNA Technologies) in Supplementary Table 3.
Sequences coding for M. musculus SPO11 and TOP6BL were codon optimized for expression in Sf9 cells and synthesized as gBlocks. The SPO11 gBlock was cloned into pFastBac1-MBP to yield pCCB630 (MBP–SPO11), and the TOP6BL gBlock was cloned into pFastBac1-HisFlag to yield pCCB628 (His–mTOP6BL–Flag).
The SPO11-Y137F/Y138F mutant was generated by QuikChange mutagenesis of pCCB630 using primers cb886 and cb887 to yield pCCB642 (MBP–SPO11-YFYF). Other SPO11 active site mutants were generated by inverse PCR and self-ligation using template pCCB630. Primers and the resulting plasmids are as follows: SPO11-Y137F (primers cb1577 and cb1578, plasmid pCCB1084), SPO11-Y138F (primers cb1579 and cb1580, plasmid pCCB1085) and SPO11-E224A (primers cb1581 and cb1582, plasmid pCCB1086).
Expression and purification of recombinant proteins
Viruses were produced using the Bac-to-Bac Baculovirus Expression System (Invitrogen) according to the manufacturer’s instructions. We infected 4 × 109 Spodoptera frugiperda Sf9 cells (Gibco, Thermo Fisher) with viruses at a multiplicity of infection of 2. Expression of MBP–SPO11 used viruses generated from pCCB630, and MBP and His–Flag tagged SPO11–TOP6BL complexes used viruses generated from pCCB630 and pCCB628. Following 72 h of infection, cells were collected, washed with PBS, frozen in dry ice and kept at −80 °C until use. All purification steps were carried out at 0–4 °C. Cell pellets were resuspended in 80 ml of lysis buffer (50 mM HEPES-NaOH pH 6.8, 1 mM DTT, 2 mM EDTA and protease inhibitor cocktail (Sigma-Aldrich, catalogue no. P8340) diluted 1:800, supplemented with 4 µM leupeptin, 5.8 µM pepstatin, 6.6 µM chymostatin and 1 mM phenylmethanesulfonyl fluoride), pooled in a beaker and then mixed slowly with a stir bar for 20 min. Ice-cold glycerol (10%) and 1 M NaCl were added to the cell lysate, which was then centrifuged at 43,000g for 25 min. The cleared extract was loaded onto 2 ml of pre-equilibrated amylose resin (NEB). The column was washed extensively with amylose buffer (25 mM HEPES-NaOH pH 6.8, 1 M NaCl, 5% glycerol, 1 mM DTT and 2 mM EDTA) and eluted with buffer containing 10 mM maltose. Fractions containing protein were loaded on a HiLoad 16/600 Superdex 200 pg column pre-equilibrated with buffer containing 25 mM HEPES 6.8, 100 mM NaCl, 2 mM DTT and 5 mM EDTA. The peak was collected and diluted twofold in buffer without salt, loaded on a Capto HiRes cation exchange column and eluted with a 0.1–0.5 M NaCl gradient. Fractions containing purified proteins were pooled, and aliquots flash-frozen in liquid nitrogen and stored at −80 °C.
For the MBP and His–Flag tagged SPO11–TOP6BL complexes, Sf9 cells were lysed by raising salt concentration to 500 mM. Cleared extract was incubated for 20 min with 2 ml of pre-equilibrated Ni-NTA resin (Thermo Scientific) and washed extensively with a buffer containing 25 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 0.1 mM DTT and 40 mM imidazole. The complex was eluted with buffer containing 500 mM imidazole and loaded onto 2 ml of equilibrated amylose resin. The resin was washed with 25 mM HEPES 7.5, 500 mM NaCl, 10% glycerol, 1 mM DTT and 2 mM EDTA, and the protein eluted with buffer containing 10 mM maltose. Fractions containing protein were loaded on a Superdex 200 column equilibrated in 25 mM HEPES pH 7.5, 400 mM NaCl, 10% glycerol, 2 mM DTT and 5 mM EDTA. The peak was collected, concentrated using 10-kDa Amicon centrifugal filters (Millipore), aliquoted, flash-frozen in liquid nitrogen and stored at −80 °C.
SEC–MALS
Light-scattering data were collected using a Superdex 200 increase 10/300 GL SEC column connected to the AKTA Pure Chromatography System (Cytiva). The elution from SEC was monitored using a differential refractometer (Optilab, Wyatt) and a static, dynamic, multiangle laser light-scattering detector (miniDAWN, Wyatt). The SEC–ultraviolet/light-scattering/refractive index system was equilibrated in buffer containing 25 mM HEPES-NaOH pH 7.5, 500 mM NaCl, 10% glycerol, 5 mM EDTA and 2 mM DTT, at a flow rate of 0.3 ml min−1. Average molecular mass was determined across the entire elution profile at intervals of 0.5 s from static light-scattering measurement using ASTRA software (Wyatt).
Plasmid cleavage assay
Cleavage reactions (20 µl) were typically carried out with 250 nM MBP–SPO11 and 5 ng µl−1 pUC19-derived 3-kb plasmid (pCCB959) in buffer containing 25 mM Tris pH 7.5, 5% glycerol, 50 mM NaCl, 1 mM DTT, 0.1 mg ml−1 bovine sreum albumin (BSA), 5 mM MgCl2 and 1.5 mM MnCl2, unless stated otherwise. Reactions were incubated for 2 h at 37 °C, stopped with 50 mM EDTA and 1% SDS and treated with 0.2 mg ml−1 proteinase K for 15–30 min at 55 °C. DNA was separated on a 1% TBE-agarose gel and stained using SYBR Gold.
For phenol–chloroform partitioning of cleavage products, cleavage reactions were stopped in the presence or absence of proteinase K. Following 20-min incubation at 55 °C, samples were mixed with an equal volume of phenol–chloroform–isoamyl alcohol and centrifuged for 5 min at 13,000 rpm. The organic phase and interphase were back-extracted twice with 100 mM Tris-HCl pH 8.0, 1 mM EDTA and 200 mM NaCl; the organic and aqueous phases were ethanol precipitated. DNA was resuspended in buffer containing 30 mM Tris-HCl pH 8.5, 1 mM EDTA, 100 mM NaCl and 0.2 mg ml−1 proteinase K, with incubation for 1 h at 55 °C. DNA was again ethanol precipitated, resuspended in TE buffer, separated on 1% TBE-agarose gel and stained using SYBR Gold. Data were quantified using ImageJ2 and plotted with GraphPad Prism 9.
For the analysis of DNA cleavage with linear substrates containing zero, one, three or six copies of the Widom 601 sequence, fragments containing one, three or six copies of Widom 601 were cloned into the multiple cloning site of pUC19 to yield pCCB1106, pCCB1107 and pCCB1108, respectively. The plasmids were PCR amplified with primers pl68 and vg001 (containing a 5′ 6-carboxyfluorescein dye) to yield linear substrates for the reaction. Cleavage reactions were performed under standard conditions with 500 nM MBP–SPO11 and 25 ng µl−1 linear fluorescent substrate. DNA was separated on 1% TBE-agarose gel and visualized using a Typhoon scanner (Cytiva).
Nicked DNA substrates were prepared by treatment of pOC157 with Nb.BrsDI, followed by phenol extraction and ethanol precipitation.
Detection of fluorescent SPO11–DNA covalent complexes
Substrates were assembled by annealing primers dd77 and cb100, or cb1593 and cb100, to produce 80-bp duplex DNA with a 6-FAM fluorophore located in 5′ or 3′, respectively. Oligos were mixed in equimolar concentrations (10 μM) in 100 mM NaCl, 10 mM Tris-HCl pH 8.0 and 1 mM EDTA, heated and slowly cooled on a PCR thermocycler (3 min at 98 °C, 1 h at 75 °C, 1 h at 65 °C, 30 min at 37 °C and 10 min at 25 °C).
Cleavage reactions (20 µl) contained 1 µM MBP–SPO11 and 0.5 µM fluorescent substrate in buffer, containing 25 mM Tris pH 7.5, 5% glycerol, 10% DMSO, 40 mM NaCl, 1 mM DTT, 5 mM MgCl2 and 1.5 mM MnCl2. Reactions were incubated for 2 h at 37 °C, stopped with 1× Leammli buffer and separated by SDS–PAGE. Fluorescent gel was scanned using a Typhoon scanner (Cytiva), and proteins stained with Coomassie blue.
Sequencing gel analysis of SPO11 cleavage sites
Oligonucleotides cb95 and cb100 were first purified on 10% polyacrylamide-urea gels. For each oligo, 5 pmol was 5′-end labelled with [γ-32P]ATP and T4 polynucleotide kinase (NEB). The labelled oligo was mixed in equimolar concentrations with the unlabelled reverse complement and annealed by heating at 100 °C in a water bath, followed by slow cooling. Labelled substrates were then purified by native PAGE.
Cleavage reactions (20 µl) contained 500 nM MBP–SPO11, 1 nM radioactive substrate and 2.5 nM (100 ng) plasmid DNA in buffer containing 25 mM Tris pH 7.5, 5% glycerol, 0.1 mg ml−1 BSA, 50 mM NaCl, 1 mM DTT, 5 mM MgCl2 and 1.5 mM MnCl2. Reactions were incubated for 2 h at 37 °C then stopped with 50 mM EDTA and 1% SDS. Markers were generated by partial digestion of substrate using either the indicated restriction enzymes or DNase I. DNA was ethanol precipitated and separated on 10% TBE-UREA sequencing gel; the gel was then dried and developed by autoradiography.
Gel shift assays
The hairpin substrate was assembled by self-annealing of primer cb957. The substrate was 5′ end labelled with [γ-32P]ATP (Revvity) and T4 polynucleotide kinase (NEB) and purified by native PAGE. Binding reactions (20 µl) were carried out in 25 mM Tris-HCl pH 7.5, 7.5% glycerol, 50 mM NaCl, 2 mM DTT, 5 mM EDTA, 1 mg ml−1 BSA and the indicated amounts of protein complexes. Gel shift assays with radioactive substrates contained 0.1 nM DNA. Reactions were incubated for 30 min at 37 °C and separated on 7% TAE-polyacrylamide/bis (37.5:1) gel at 200 V for 2 h. Gels were dried, exposed to autoradiography plates and demonstrated by phosphorimaging. Gel shift assays with plasmid substrates contained 100 ng DNA. Reactions were incubated for 30 min at 37 °C and separated on 1% TAE-agarose gels at 60 V for 2 h. Gels were stained with SYBR Gold and scanned using a Typhoon scanner (Cytiva).
Statistics and reproducibility
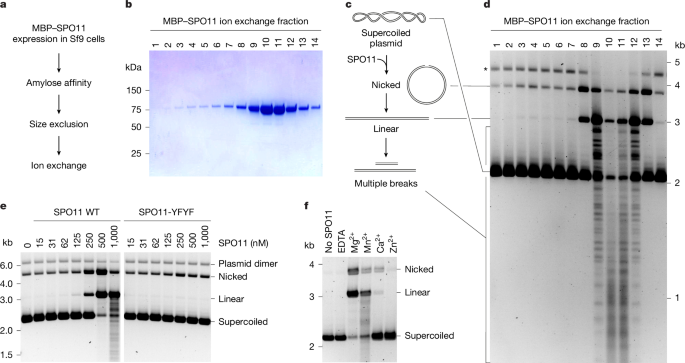
Sample numbers in quantifications are indicated in the figure legends. Gels shown in the article are representative images. In Fig. 1b,d, ion exchange fractions of purified proteins were analysed by gel electrophoresis and assayed for DNA-cleavage activity at least twice. In Fig. 1e,f, the importance of active site tyrosines and metal ions was confirmed more than three times. In Fig. 2b–e, experiments were performed at least twice, with similar results. In Fig. 3a–c, cleavage sites were mapped on the various substrates at least twice, with similar results. In Fig. 4b,c,e, experiments were reproduced at least twice. In Fig. 1e, DNA-binding activities of the complexes were compared twice, with similar results. In Extended Data Fig. 1a,b, experiments were performed once but the conclusions were confirmed multiple times independently. In Extended Data Fig. 2a, the experiment was performed once. In Extended Data Fig. 3a, the quantification shows a single experiment but the observation was reproduced at least twice under different conditions. In Extended Data Fig. 4d, the observation was reproduced more than three times. In Extended Data Fig. 5a,b, the observations were reproduced twice under different conditions. In Extended Data Fig. 6a,b, quantifications show a single experiment but the observation was reproduced at least twice under different conditions. In Extended Data Fig. 6f,g, observations were reproduced at least twice under different conditions. In Extended Data Fig. 7a–c, observations were reproduced at least twice. No statistical methods were used to predetermine sample size. Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.