Mice
C57BL/6J mice 8–12 weeks of age were obtained from the Jackson Laboratory (JAX: 000664). Ccr2-KO (strain number 004999), Sarm1-KO (strain number 018069), CCL2–RFP (strain number 016849) and Lgals3-KO (strain number 006338) mice were obtained from the Jackson Laboratory. All animal experiments and procedures were conducted according to the institutional animal care and safety guidelines at Boston Children’s Hospital. All mice in this study were kept on a 12 h light cycle, at 21–23 °C, with 30–50% humidity.
Diet
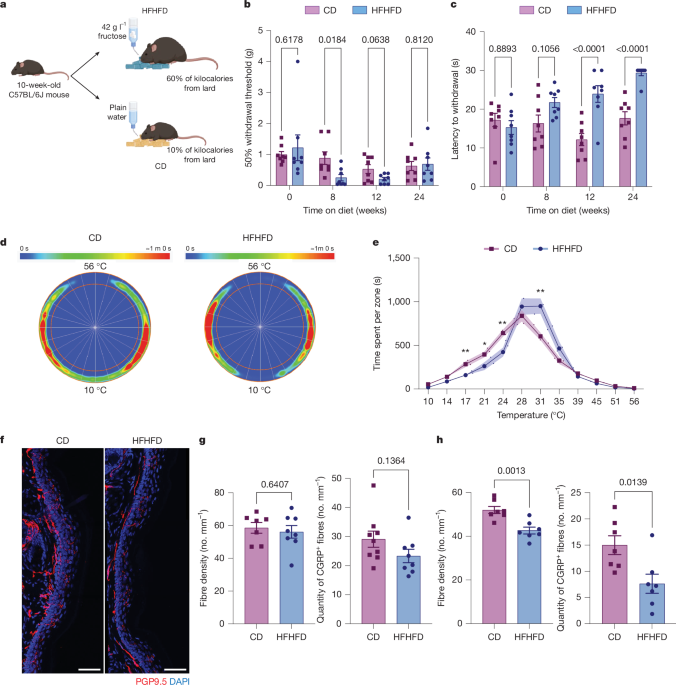
A high-fat diet containing 60% fat from lard was obtained from Research Diets (D12492), along with a matched sucrose CD (D12450J). Diet experiments were started at 10–12 weeks of age. Whole cages of mice were randomly assigned to receive the CD or HFHFD. Mice that were given the HFHFD were also given water bottles with 42 g l−1 d-(−)-fructose in autoclaved water, and those given CD were given plain autoclaved water. Mouse food and water levels were monitored three times per week and replenished as needed throughout the entirety of their diet regimen.
Glucose tolerance test
Mice were fasted overnight, and baseline blood glucose was measured from a tail nick using a One Touch Ultra 2 glucose monitoring system. Mice were then injected with 5 μl g−1 body weight of 0.2 g ml−1 glucose i.p., and blood glucose was measured at 15, 30, 60 and 120 min.
HbA1c assay
Mice were fasted for 4 h, and whole blood was collected using cardiac puncture with EDTA and stored at −20 °C. The Mouse Hemoglobin A1c assay kit (Crystal Chem, catalogue number 80310) was used to measure mouse glycated haemoglobin levels following the manufacturer’s protocol. In brief, blood samples were lysed using the provided lysis buffer and the absorbance was measured at 700 nm twice following two enzymatic protease digestions. The provided calibrators were used to calculate the percentage of glycated HbA1c in each sample.
Insulin enzyme-linked immunosorbent assay
Mice were fasted for 4 h, and whole blood was collected using cardiac puncture with EDTA. Blood was then centrifuged at 4 °C for 20 min at 2,000g to collect plasma, which was then frozen at −20 °C until the assay was carried out. Insulin levels in each sample were measured using the Ultrasensitive Mouse Insulin ELISA kit (Crystal Chem, catalogue number 90080) following the manufacturer’s protocol for the wide-range assay. In brief, samples and standard solutions were incubated in the pre-coated enzyme-linked immunosorbent assay wells provided. The plate was then washed and incubated in an anti-insulin-conjugated solution followed by washes and substrate solution incubation. Finally, the reaction was stopped, and the absorbance was measured at 450 nm and 630 nm and calculated using the standard curve.
Behaviour
von Frey
Mice were habituated to von Frey chambers and mesh bottoms for 2 days, 1 h each day, before testing. On the day of the test, mice were habituated for 1 h before the test. Mice were placed in behaviour chambers in random order at every timepoint as a mean of blinding investigators, and there was no consistent placement for each mouse. The up–down method was used to determine the mice’s 50% withdrawal threshold58. Behavioural experiments to compare CD and HFHFD were otherwise difficult to conduct in a blinded fashion, as mouse weights and faeces colours were obvious; however, these were conducted by at least two independent investigators to validate the findings. All subsequent experiments with genetic or pharmacological manipulations were conducted in a blinded fashion (as was the Hargreaves test).
Hargreaves
Mice were habituated to the Hargreaves apparatus (IITC number 390G) consisting of a glass floor heated to 30 °C and a Plexiglass chamber for 2 days before testing, 30 min each day. On the day of the assessment, mice were habituated for 1 h before the test. A focused heat light source was shined on the plantar surface of the left paw of the mice, and a ramping heat stimulus was therefore applied until paw withdrawal was recorded, with a time limit of 30 s. Readings were averaged from three trials per mouse.
Thermal gradient ring
Mice were placed in the thermal gradient ring (Ugo Basile, 35550), which was set to 10 °C on one side and 56 °C on the other side. Their location and movement and the temperature of the plates were recorded for an hour. All mice in a single experiment were recorded using one apparatus. Using the ANY MAZE software, the time spent per zone and distance travelled were recorded for each mouse.
Skin immunohistochemistry and IENFD analysis
A 2-mm punch from the hind paw plantar skin of mice was collected and fixed in Zamboni Fixative (Newcomer, number 1459A) overnight at 4 °C on a piece of filter paper to flatten the tissue. After fixation, skin samples were kept in 30% sucrose for at least 48 h in 4 °C. Skin samples were then frozen in OCT, cryosectioned at 30 μm thickness and mounted onto SuperFrost Plus microscope slides. For immunostaining, slides were thawed for 1 h at room temperature and then washed in PBS for 15 min following three 10-min 1% Triton-X washes. Slides were incubated in blocking buffer for 2 h at room temperature (10% donkey serum, 0.4% Triton-X, 0.05% Tween 20, 1% BSA) and then incubated in primary antibody overnight at 4 °C (rabbit anti-PGP9.5 Abcam catalogue number ab108986, 1:200). The next day, slides were washed three times with PBS and incubated in secondary antibody for 2 h at room temperature (donkey anti-rabbit Cy3 Jackson ImmunoResearch 711-165-152 1:500). Slides were then washed three times with PBS and mounted with Prolong Antifade DAPI medium (Invitrogen, number P36935). Confocal images were taken on a Leica SP8 microscope with a ×63 oil objective. A z-stack spanning the 30 μm thickness was taken and four adjacent images were tiled. Using FIJI (ImageJ), analysis to determine IENFD was conducted by an investigator blinded to the treatment groups. Maximum-intensity projection was taken, and the number of fibres crossing the dermo-epidermal boundary was counted and divided by the length of the epidermal layer in the image to obtain an IENFD value for that image following previously described rules59. At least three images of non-consecutive sections were averaged per mouse to obtain the IENFD value for that mouse.
Sciatic nerve tissue collection and processing
Sciatic nerves were dissected and collected in ice-cold 1% BSA in RPMI. For digestion, tissues were placed in collagenase A (Roche, 5 mg ml−1) and Dispase II (Roche, 1 mg ml−1) and were minced before being moved to a thermomixer at 37 °C for 1 h at 1,000 r.p.m. Samples were then filtered using a 70-μm filter, washed in FACS buffer (2% fetal bovine serum (FBS), 2 mM EDTA, PBS) and then washed with 1× PBS to obtain single-cell suspensions.
Skin tissue collection and processing
Skin punch biopsies were dissected and collected in ice-cold 1% BSA in RPMI. Tissues were minced and Liberase (Sigma-Aldrich, catalogue number 05401119001) was added at a final concentration of 1 mg ml−1, and tubes were placed on a thermomixer at 37 °C for 1.5 h at 1,000 r.p.m. Samples were then filtered using a 70-μm filter, washed in FACS buffer (2% FBS, 2 mM EDTA, PBS) and then washed with 1× PBS to obtain single-cell suspensions.
Dorsal root ganglion collection and processing
Lumbar dorsal root ganglia were collected in ice-cold Dulbecco’s modified Eagle medium (DMEM) containing FBS plus penicillin and streptomycin. Dorsal root ganglia were digested in collagenase A (Roche, 5 mg ml−1) and Dispase II (Roche, 1 mg ml−1) for 70 min. Digested dorsal root ganglia were triturated using large-, medium- and small-sized polished glass pipettes in DMEM containing DNase. The cells were resuspended in DMEM containing FBS plus penicillin and streptomycin and were overlaid on 10% BSA solution. The bilayer was centrifuged for 12 min at 1,000g at reduced acceleration and deceleration. The top two layers were discarded, and the cell pellet was resuspended in FACS buffer (2% FBS, 2 mM EDTA, PBS) and then washed with 1× PBS to obtain single-cell suspensions.
Flow cytometry
Cells were then incubated for 10 min at 4 °C in Fc Block (Tonbo Biosciences, catalogue number 70-0161-U500, 1:2,000) and then washed with FACS buffer. Samples were then stained for 30 min with antibodies to mouse CD45–ef780 (Thermo Fisher Scientific, clone 30-11, catalogue number 47-0451-82, 1:400), CD11B–FITC (Thermo Fisher Scientific, clone M1/70, catalogue number 11-0112-82, 1:400), CD64–PE-594 (BioLegend, clone X54-5/7.1, catalogue number 139319, 1:400), LY6C–BV711 (BioLegend, clone HK1.4, catalogue number 128037, 1:1,000), LY6G–PE (BioLegend, clone 1A8, catalogue number 127607, 1:800), CD3ε–PE (BioLegend, clone 145-2C11, catalogue number 100307, 1:400) for 30 min on ice and then washed with FACS buffer twice and resuspended with 3 μM DAPI before being run on an LSR Fortessa Cytometer using BDFACS Diva Software and analysis using FlowJo.
Preparation of immune cells for single-cell sequencing
Single-cell suspensions collected as described above from two mice (four sciatic nerves) were pooled in each sample. Samples were resuspended in FACS buffer (2% FBS, 2 mM EDTA, PBS) and then incubated for 10 min at 4 °C in Fc Block (Tonbo Biosciences catalogue number 70-0161-U500, 1:2,000) and washed with FACS buffer. Cells were then stained for 30 min in anti-CD45 conjugated APC–ef780 antibody (Thermo Fisher Scientific, clone 30-11, catalogue number 47-0451-82, 1:200) in FACS buffer and then washed twice in PBS with 0.5% BSA and no EDTA. Cells were then resuspended with 3 μM DAPI in PBS with 0.5% BSA before live CD45+ cells were sorted on a BD FACS Aria sorter using BDFACS Diva Software and processed for 10× sequencing following a 10× protocol.
Barcoding and library preparation
The Chromium Next GEM Single Cell 3′ Reagent kit v3.1 (Dual Index) was used. Barcoding and library preparation were performed following the manufacturer’s protocols. In brief, to generate single-cell gel-bead-in-emulsion solution, sorted cells were resuspended in a final volume of 40 μl and were loaded on a Next GEM Chip G (10X Genomics) and processed with the 10X Genomics Chromium Controller. Reverse transcription was performed as follows: 53 °C for 45 min and 85 °C for 5 min in a thermocycler. Next, first-strand cDNA was cleaned with DynaBeads MyOne SILANE (10X Genomics, 2000048). The amplified cDNAs, intermediate products and final libraries were prepared and cleaned with the SPRIselect Regent kit (Beckman Coulter, B23318), and examined on a High-Sensitivity DNA Chip with the Bioanalyzer system (Agilent). Ten-microlitre aliquots of cDNAs were use for library preparation following the manufacturer’s instructions (10X Genomics). The final libraries were examined on High-Sensitivity DNA Tape with TapeStation (Agilent) before pooling for sequencing. A small aliquot of each library was used for quality control to determine fragment size distribution and DNA concentration using a bioanalyser. Libraries were pooled for sequencing with a NovaSeq 6000 (Illumina) at an estimated depth of 113,000 reads per cell in the nerve and 65,000 reads per cell in the skin. Sequencing reads were aligned to the mouse reference transcriptome (mm10, version 2020-A) using CellRanger (v7.0.1).
Single-cell RNA-sequencing data analysis
Raw single-cell RNA-sequencing data were processed using 10X Genomics CellRanger software version 7.0.01. The CellRanger mkfastq function was used for de-multiplexing and generating FASTQ files from raw BCL. The CellRanger count function with default settings was used with the mm10 reference supplied by 10X Genomics, to align reads and generate single-cell feature counts. Ambient RNA contamination was removed from each sample using CellBender6 (remove-background, default parameters). Seurat version 5.0.1 implemented in R version 4.3.2 was used for downstream analysis. Cells were excluded if they had fewer than 500 features, or fewer than 250 genes, or the mitochondrial content was more than 20%. Two CD and two HFHFD samples were integrated and normalized following a previously described Seurat SCTransform+ CCA integration pipeline60. The mitochondrial mapping percentage was regressed out during the SCTransform normalization step. PCA was performed and the top 40 principal components were used for downstream analysis. A K-nearest-neighbour graph was produced using Euclidean distances. The Louvain algorithm was used with the resolution set to 0.4 to group cells together. Nonlinear dimensional reduction was carried out using UMAP. The FindAllMarkers function was used to determine marker genes for each cluster and clusters were identified on the basis of canonical markers previously published in the literature5,13,29. Differentially expressed genes were determined using the FindMarkers function that performs differential expression testing based on the non-parametric Wilcoxon rank sum test.
Integration with macrophages after nerve crush dataset
All clusters expressing Csf1r were subset into a new dataset and raw data file from ref. 5 were downloaded. Samples from two CD mice and two HFHFD mice were merged using Seurat with Ydens’ naive, day 1 post-crush and day 5 post-crush samples. Data were filtered according to the same filters above and integrated using SCTransform as described above. The dataset was then re-clustered with 0.4 resolution, and FindAllMarkers was used to identify cell types.
Sciatic nerve immunohistochemistry
Mice were anaesthetized after 12 weeks of CD or HFHFD feeding using 200 mg kg−1 pentobarbital and perfused transcardially using PBS. Sciatic nerves were collected in 4% paraformaldehyde and left in 4 °C overnight and then moved to 30% sucrose solution. Nerves were then frozen in OCT, cryosectioned as 50-μm cross-sections and mounted onto SuperFrost Plus microscope slides. For immunostaining, slides were thawed for 1 h at room temperature and then washed in PBS for 15 min following three 10-min 1% Triton-X washes. Slides were incubated in blocking buffer for 2 h at room temperature (10% donkey serum, 0.4% Triton-X, 0.05% Tween 20, 1% BSA) and then incubated in primary antibody overnight at 4 °C: rat anti-F4/80 (Bio-Rad, catalogue number MCA497GA, 1:250), goat anti-collagen type IV (Sigma-Aldrich, catalogue number AB769, 1:500), goat anti-CD31 (Novus Biologicals, catalogue number AF3628, 1:25). The next day, slides were washed three times with PBS and incubated in secondary antibody for 2 h at room temperature: donkey anti-rat 488 (Abcam, catalogue number ab150153, 1:500), donkey anti-goat 647 (Invitrogen, catalogue number A21447, 1:500). Slides were then washed three times with PBS and mounted with Prolong Antifade DAPI medium (Invitrogen, catalogue number P36935). Confocal images were taken on a Leica SP8 microscope with a ×20 air objective. A z-stack spanning the whole thickness was taken. A maximum-intensity projection was generated using ImageJ, and the area within the epineurium (collagen staining) was measured. The number of F4/80+ macrophages associated with a nucleus was counted and divided by the area of the endoneurial space in each image.
Nerve electron microscopy
Sciatic nerves were collected in 2.5% glutaraldehyde, 2.5% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.5) and fixed in the same solution overnight. Small pieces (1–2 mm) of fixed tissue were washed in 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide (OsO4) and 1.5% potassium ferrocyanide (KFeCN6) for 1 h, washed in water twice, then washed once in 50 mM maleate buffer pH 5.15 (MB) and incubated in 1% uranyl acetate in MB for 1 h followed by one wash in MB, two washes in water and subsequent dehydration in grades of alcohol (10 min each; 50%, 70%, 90%, 2 × 10 min 100%). The samples were then placed in propyleneoxide for 1 h and infiltrated overnight in a 1:1 mixture of propyleneoxide and TAAB Epon (TAAB Laboratories Equipment). The next day, samples were embedded in TAAB Epon and polymerized at 60 °C for 48 h. Ultrathin sections were cut on a Reichert Ultracut-S microtome, picked up on copper grids, stained with lead citrate and examined in a JEOL 1200EX Transmission electron microscope or a TecnaiG2 Spirit BioTWIN. Images were recorded with an AMT 2k CCD (charge-coupled device) camera. Electron microscopy imaging, consultation and services were performed at the Electron Microscopy Facility at Harvard Medical School. g ratios and percentages of myelinated axons were calculated from the EM images using MyelTracer as described before61.
Mouse injections
The CCR2 and CCR5 inhibitor CVC (Selleckchem, catalogue number S8512) or dimethylsulfoxide was diluted in corn oil and injected i.p. at 20 mg kg−1 three times per week starting 8 weeks on the HFHFD until 12 weeks. Mice were chosen randomly from each cage. Mouse weights were recorded weekly, and two mice in each group that lost weight as a result of the drug treatment were excluded from the study before proceeding with behavioural testing because this counteracted the weight gain expected from HFHFD feeding. For behavioural testing, the person injecting mice or placing mice in behaviour chambers was different from the person investigating the behaviour, who was therefore fully blinded.
Sciatic nerve crush surgery
Mice were anaesthetized by administering 2.5% isoflurane. A sciatic nerve crush was performed by exposing the left sciatic nerve at the mid-thigh level and crushing it with a haemostat at the second stop for 30 s. The surgical incision was then closed using silk sutures and the mouse was monitored every day after the surgery.
RT–qPCR
Mouse sciatic nerves were collected in RNAlater and stored at 4 °C overnight before long-term storage at −80 °C. To isolate RNA, nerves were moved from RNAlater to RLT buffer (Qiagen) and minced. A handheld homogenizer was then used to homogenize the nerves in RLT buffer, and an equal volume of 70% ethanol was then added to the tube. RNA was isolated following the Qiagen mini kit protocol. cDNA was synthesized from the resulting RNA using the cDNA VILO kit (Invitrogen, catalogue number 11754050). Relative gene expression was determined using gene-specific primers (PrimerBank) and SYBR Green master mix (Life Technologies) on a 7500 Fast Real-time PCR system (Applied Biosystems). Expression levels were normalized to Hprt levels using the 2−ΔCt method. The following primer sequences were used—Hprt forward: 5′-CAGTCCCAGCGTCGTGATTA-3′, Hprt reverse: 5′-TGGCCTCCCATCTCCTTCAT-3′; Ccl2 forward: 5′-GATGCAGTTAACGCCCCACT-3′, Ccl2 reverse: 5′-GAGCTTGGTGACAAAAACTACAGC-3′, Lgals3 forward: 5′-TAATCAGGTGAGCGGCACAG-3′, Lgals3 reverse: 5′-ATAGGGCACCGTCAGTGGTC-3′, Trem2 forward: 5′-ATGGGACCTCTCCACCAGTT-3′, Trem2 reverse: 5′-CACAGGATGAAACCTGCCTGG-3′.
Statistical analysis
Sample size for behavioural experiments was based on previous laboratory data and data in the literature62,63,64. Sample size for the single-cell RNA-sequencing experiment was based on common practice at n = 2 for single-cell transcriptomics. We also utilized an available tool to estimate sample size65 using our knowledge of the frequency of rare populations (for example, neutrophils in healthy tissue), and determined that the number of cells in our analyses was above that needed for 95% probability of success at detecting rare populations. Statistical analysis was performed using GraphPad Prism software. For each experiment of its kind, we conducted a normality test using the Shapiro–Wilk test, and they all passed the normality test; therefore, we assumed normality for subsequent tests from the same datasets. This was the case for body weight, von Frey, Hargreaves, flow cytometry, IENFD, fasting insulin, glucose tolerance test, HbA1c and nerve immunohistochemistry data. For statistical comparison between groups, two-group comparisons were made using two-tailed unpaired Student’s t-test. For comparisons of two groups over time, two-way ANOVA with appropriate multiple comparisons tests was used, and 0.05 was set as the threshold for significance. Plots from behaviour assays, immunohistochemistry and flow cytometry are representative of at least two independent repetitions. All error bars plotted represent s.e.m.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.