Cell Lines
MDA-MB-231, NCI-H1299, PC-3, MCF7, HCC38, UMUC3, NCI-H1975, PANC-1, NCI-H460, MDA-MB-468, A549, NCI-H1437, SNU-44948, U118-MG, CAPAN-1, DAOY, MIAPACA2, NCI-H838, NCI-H1755, LN18, NCI-H747, SW-837, T84, COLO205, COLO201, RKO, DLD-1, SW48, HEMa, TFF-1, ARPE-19, MCF-10A and HEK-293 cells were purchased from the American Type Culture Collection (ATCC). KYSE-450, MFE-319 and COLO678 cells were purchased from Leibniz Institute DSMZ. U87-MG was acquired from Ludwig Institute. LK-2 was acquired from the Japanese Collection of Research Biosources (JCRB), HT-29 was acquired from the National Institute of Health (NIH) and NCI-H322, HT115 and HT55 were acquired from the European Collection of Authenticated Cell Cultures (ECACC). All cell lines were confirmed for identity by short tandem repeat profiling and confirmed to be negative for mycoplasma. All cell lines were cultured in DMEM or RPMI (Gibco) supplemented with 10% heat inactivated FBS (Gibco).
Antibodies
The following primary antibodies were used to detect protein abundance in Simple Western assays: PELO (ProteinTech, 10582-1-AP), HBS1L (ProteinTech, 10359-1-AP), SKIV2L (ProteinTech, 11462-1-AP), TTC37 (Abcam, ab122421), WDR61 (Invitrogen, PA5-59052), FOCAD (Invitrogen, PA5-63051), AVEN (ProSci, 2413), ABCE1 (Abcam, ab185548), JNK (Cell Signaling, 4672), pJNK (Cell Signaling, 4668), p38 (Cell Signaling, 9212), phospho-p38 (Cell Signaling, 9211), EIF2S1 (Cell Signaling, 5324), phospho-EIF2S1 (Abcam, ab32157), IRE1α (Cell Signaling, 3294), pIRE1α (Novus Biologicals, NB100-2323), PERK (Cell Signaling, 3192), phospho-PERK (Cell Signaling, 3179), CHOP (ThermoFisher Scientific, MA1-250), JUN (Cell Signaling, 9165), p21 (Cell Signaling, 2947), PARP (Cell Signaling, 3898), HSP90 (Cell Signaling, 4877), GAPDH (Cell Signaling, 2118), HiBIT (Promega, N7200). Antibodies were diluted at 1:100 except for HSP90 (1:500) and GAPDH (1:3,000). High-content antibodies as follows: rabbit anti-IRE1α (Invitrogen PA5-20189), dilution 1:500. Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluo 568 (Thermofisher A-11011), dilution 1:500.
In vivo studies
All animal studies were conducted in compliance with AbbVie’s Institutional Animal Care and Use Committee and the National Institutes of Health Guide for Care and Use of Laboratory Animals guidelines in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
In vivo CRISPR screening
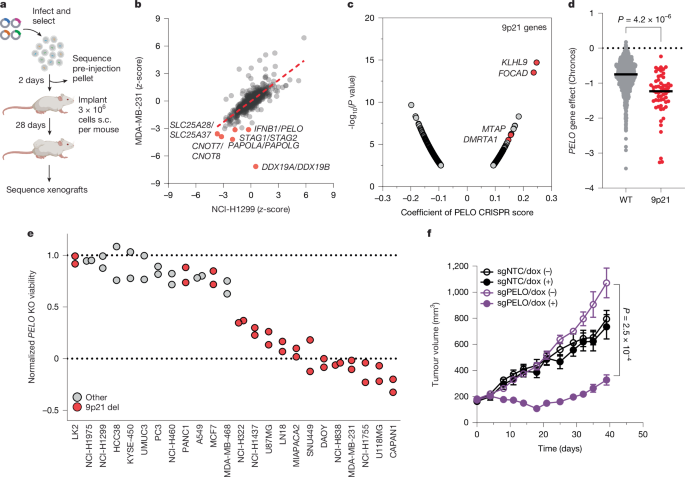
MDA-MB-231 and NCI-H1299 cells were engineered to express enhanced Cas12a (enCas12a)49, as described50. Each line was transduced for 24 h with a Cas12a customized sgRNA library, as described11 at a MOI of 0.3, maintaining continuous 1,000× coverage (3,000,000 cells per replicate) through selection. Following selection with 2 µg ml−1 puromycin for 48 h, cells were washed briefly in media absent of antibiotics and inoculated into one flank (NCI-H1299) or into the mammary fat pad (MDA-MB-231) of 10 NOD scid gamma (NSG) mice per arm. Tumours were collected when measured at above 500 mm3 by calliper. DNA extraction, PCR and next-generation sequencing were performed as previously described51 at 1,000× coverage using 120-nt single-end reads on a NextSeq 500 (Illumina). All cells and lentivirus used were tested for rodent pathogens and mycoplasma by IDEXX Bioanalytics.
CRISPR and siRNA validation of PELO and HBS1L dependency
For CRISPR–Cas9 knockout, unless otherwise noted, negative control guides were targeted to the non-essential gene B2M. Positive control guides include those designed to target essential genes (RAN and RPA3) and the common essential gene (ABCE1). A full list of sgRNA sequences used can be found in Supplementary Table 1. To perform CRISPR gene editing across cancer cell lines, we utilized electroporation of ribonucleoprotein complexes (RNPs). In short, functional sgRNA duplexes were created by hybridizing Alt-R CRISPR–Cas9 CRISPR RNA (IDT) with Alt-R CRISPR–Cas9 trans-activating CRISPR RNA (IDT). These sgRNA complexes were incubated with Alt-R Streptococcus pyogenes Cas9 Nuclease V3 (IDT) and electroporation enhancer (IDT) to create RNPs, which were then electroporated into the cell using the Invitrogen’s Neon NxT Electroporation system according to the manufacturer’s protocol.
To generate doxycycline-inducible models, MIAPACA2 cells were stably derived via lentiviral infection of pXPR_BRD10152 (Broad Institute). After Cas9 infection and selection with 20 µg ml−1 blasticidin (Gibco), Cas9 activity was measured and deemed sufficient if at least 80% knockout of a known control gene was achieved. All Cas9 models described in this study are polyclonal unless otherwise mentioned. After confirmation of Cas9 activity, doxycycline-inducible NTC, PELO or HBS1L sgRNA constructs (Cellecta) were stably integrated by 2 µg ml−1 puromycin selection. Generated doxycycline-inducible Cas9–sgRNA cells were cultured in RPMI supplemented with 10% tetracycline (tet) free FBS (Takara), treated with 2 µg ml−1 doxycycline (Sigma) to induce knockout and validated by western blot.
For small interfering RNA (siRNA) validation of HAP1 (Horizon Discovery) dependency, cells with or without SKIV2L knockout were plated at 1,000 cells per well in 96-well plates and reverse transfected with 10 nM NTC (Dharmacon, J-008702-06), PELO (Dharmacon, J-019068-11) or ABCE1 (Dharmacon, J-008702-06) siRNAs and Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s protocol. Knockdown efficiency was measured by Simple Western at 48 h post transfection. ON-Targetplus Non-targeting siRNA (D-001810-01-20): UGGUUUACAUGUCGACUAA; ON-Targetplus Human PELO siRNA (J-019068-11): GUGUGGUACUGGAGCGCAU; ON-Targetplus Human ABCE1 siRNA (J-008702-06): UGUCUCAGCUUGAAAUUAC.
Cell proliferation assays
Cells were plated at 1,000 cells per well (unless otherwise noted) in 96-well plates (Corning, 3903). Cell viability was assessed on day seven unless otherwise noted using CellTiter-Glo 2.0 Cell Viability Assay, according to manufacturer’s protocol (Promega, G9241), and luminescence was quantified using a PerkinElmer Envision (2104 Multilabel Reader). Cell confluency was assessed continually through Incucyte (Satorius v2022B).
Gene dependency data and informatic correlations
CRISPR dependency data can be located via the 23Q4 DepMap Public database. Two-group and continuous Pearson’s correlations of PELO CRISPR dependency data to other features were taken from the 23Q4 DepMap public portal using either mRNA expression and/or proteomic data taken from CCLE cohorts as of the 23Q4 DepMap Public release. Gene dependency, RNA, and proteomic data can be downloaded at https://depmap.org.
Conditional in vivo knockout studies
For these studies, female CB17.SCID (Charles River Laboratories) mice were used. To generate xenografts from MIAPACA2-Cas9 doxycycline-sgNTC, sgPELO or sgHBS1L cells, mice were inoculated subcutaneously into the right hind flank with 5 × 106 viable cells. The injection volume was 0.1 ml and composed of a 1:1 mixture with Matrigel (BD). Measurements of the length (L) and width (W) of the tumour were taken via electronic calliper twice weekly and the tumour volume was calculated according to the following equation: volume = L × W2/2. Mice were assigned to 2 treatment groups of 8 mice each so that the mean tumour volumes within groups were approximately 175–250 mm3. Groups were then assigned to a diet of regular chow or doxycycline chow containing 625 mg of doxycycline per kg of chow (TD.01306 Rodent Diet, Teklad). Mice were euthanized: (1) when tumour volume reached 2,000 mm3; (2) when general efficacy parameters were confidently determined; or (3) when other animal health issues were observed.
Additional cohorts of mice were entered into a biomarker study, to determine the effect of doxycycline induction on PELO and HBS1L expression in the appropriate tumour constructs. After staging into groups of 4 mice each and given access to regular chow or to doxycycline chow, tumours were excised from mice and snap frozen after 8 days, 14 days, and 21 days and analysed by Simple Western.
Exogenous expression constructs
Protein-coding expression sequences were cloned into a version of pGenLenti (lentiviral construct; Genscript) that was modified to replace the IRES sequence with an SV40 promoter to drive expression of the puromycin resistance cassette. The full-length wild type was generated from NM_015946.5 (PELO open reading frame (ORF)), NM_006620.4 (HBS1L ORF, isoform 1) or NM_006929.5 (SKIV2L ORF) and truncated fragments were synthesized and inserted using the NheI–NotI sites. All point mutants were generated using site directed mutagenesis. Sequences for the final vectors with inserts are listed in Supplementary Table 7.
Determination of protein abundance by Simple Western
Protein was extracted in CelLytic M lysis buffer (Sigma) containing cOmplete protease, phosSTOP phosphatase inhibitors (Roche), Benzonase (Sigma) and 1 mM dithiothreitol (Sigma). Sample protein concentration was measured using Pierce BCA Protein Assay reagent (ThermoFisher Scientific) and diluted to 0.5 µg µl−1 for analysis using Simple Western Jess System (Bio-Techne). Simple Western was performed according to the manufacturer’s protocol, and results were analysed using Compass software (version 6.2.0).
Isogenic model development and validation
Isogenic NCI-H1299 cell models with PELO, HBS1L, SKIV2L, TTC37, FOCAD or AVEN clonal knockout were generated via electroporation of RNPs targeting the indicated genes of interest or a negative control (B2M). To attain clonal populations, these bulk populations were then seeded in 96-well plates at a concentration of 0.25 cells per well. After 3–4 weeks, wells containing a single colony were picked and expanded. Individual clones were verified for complete knockout by Simple Western.
PELO SL-ID CRISPR screen and analysis
A library was designed for spCas9 containing 7,092 sgRNAs with 4 sgRNA constructs per gene. The lentiviral CRISPR library was produced in HEK293T cells following CRISPR & Mission Lentiviral packaging mix protocol (Sigma) and concentrated 1:10 using Lenti-X concentrator (Takara). NCI-H1299 cells engineered to express spCas9 were transduced with titrated lentiviral guide library to ensure low MOI. Cells were seeded with 5 million cells in T175 plates and infected at an MOI of approximately 0.3 at 500× representation using 5 µg ml−1 polybrene. After infection and 48 h of selection with 2 µg ml−1 puromycin, cells were either collected to assess basal library representation or maintained at 500× library representation for 14 days to allow for guide dropout and enrichment. gDNA was isolated using Nucleospin Blood kit (Takara) by manufacturer guidelines. Library generation, sequencing, and analysis of the screen were carried out as previously described51.
RNA quantification
MIAPACA2 FOCAD-dTAG cells were plated at 1 × 106 per 100 mm cell culture dish for RNA extraction. Cells were treated with 500 nM dTAGV-1 (HY-145514C, MedChemExpress) or DMSO, and samples were collected at indicated timepoints. Cells for total RNA collection were lysed on plate using RLT buffer and homogenized using QIAshredder (Qiagen) before being purified using RNeasy RNA purification kit (Qiagen) per manufacturer guidelines. Pellets for Simple Western were processed and analysed as previously described.
RNA concentration was measured by NanoVue Plus Spectrophotometer and cDNA was generated using 1 µg RNA with Superscript Vilo Master Mix (ThermoFisher Scientific) per manufacturer guidelines. Complementary DNA (cDNA) was diluted 1:100 in DEPC-treated water and two-step quantitative PCR with reverse transcription was performed in triplicate in a MicroAmp Optical 384-well reaction plate (ThermoFisher Scientific) using 3 µl diluted cDNA, 6 µl 1× Power Sybr Green PCR Master mix (ThermoFisher Scientific), 3 µl 1 µM RxNReady Primer Pools (IDT) for either FOCAD, SKIV2L, TTC37, AVEN or GAPDH, and 8 µl water. Samples were amplified using QuantStudio 6 Flex (ThermoFisher Scientific) and analysed with QuantStudio Real-Time PCR System software (version 1.3) following manufacturer guidelines. Relative quantification of gene expression was carried out with the ∆∆Ct method using housekeeping gene GAPDH Ct values and matching DMSO controls. Primers used for transcript analysis are as follows: FOCAD: ATGAAGCCAGCCTCTCCTCAGA, CTGGGTTGGTAATGGCTATCCAC; SKIV2L: GCCCAGAAACACATGACTCG, CGAAGGATCTCCGTGGTC; TTC37: TCCAGGTGATCCTGCTATCT, CTATCCTCTTTGGCTGAGTGTT; AVEN: GTCTCAGGACCTGAAATCCAAGG, GGAGGTACTTGGTTGCTCAGGT; and GAPDH: GTCTCCTCTGACTTCAACAGCG, ACCACCCTGTTGCTGTAGCCAA.
High-content analysis
Cells were washed with PBS followed by a 10-min incubation with 0.25% Triton X-100 in PBS. Plates were washed and incubated for 30 min in Click-iT EdU cocktail (Thermo C10640) followed by a second wash. Blocking solution consisting of 1% BSA in PBS was added to the plates and incubated for 30 min followed by wash. Hoechst 33342 was diluted 1:1,000 in 0.6% BSA in PBS and incubated for 30 min on plates, which were then washed and left in PBS. Plates were scanned on Thermo CX7 LZR using filter sets 405LZR_BGRFR_BGRFR (nuclei) and 647LZR_BGRFR_BGRFR (Edu). Nuclei were identified and Edu quantified using HCS Studio Target Activation.V4 algorithm.
For IRE1α foci quantification, cells were washed with PBS followed by a 15-min incubation with blocking buffer (1% BSA, 0.25% Triton X-100, 10% goat serum in PBS). Plates were washed and incubated with 1:500 diluted rabbit anti-IRE1α polyclonal antibody (Invitrogen PA5-20189) in blocking buffer at 4 °C overnight. The plates were washed and incubated with 1:500 Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (Thermofisher A-11011) and 1:1,000 Hoechst 33342 diluted in 1% BSA in PBS for 60 min, washed and left in PBS. Plates were scanned on Thermo CX7 LZR using filter sets 405LZR_BGRFR_BGRFR (nuclei) and 561 LZR_BGRFR_BGRFR (IRE1α). Nuclei were identified and IRE1α quantified using HCS Studio Target Activation.V4 algorithm.
Cell cycle and death analysis
For flow cytometry-based methods, cells were prepared for cell cycle analysis using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen) according to the manufacturer’s protocol. In brief, cells were cultured in EdU (10 μM) for 1.5–2 h prior to fixation, permeabilization, and incubation with the Click-iT reaction cocktail composed of PBS, CuSO4, Alexa Fluor 647, and buffer additive. Cells were then washed 2 times with 1× Click-iT permeabilization/wash reagent and resuspended in 500 μl FxCycle PI/RNase staining solution (Invitrogen). Cells were incubated at 4 °C for at least 15 min prior to analysis.
Cells were prepared for viability analysis using Apotracker Green (BioLegend), MitoTracker Red CMXRos (Invitrogen), and Celcein Blue AM (Invitrogen) dyes according to the manufacturers’ protocols. In brief, cells were collected, washed in FACS staining buffer (PBS + 1% BSA), and incubated in 400 nM Apotracker green for 20 min in the dark at room temperature. Cells were then washed in PBS and resuspended in 1 ml PBS containing 2 μl Calcein Blue (final concentration 1 μM) and 0.5 μl MitoTracker Red. Cells were incubated for 30 min at room temperature prior to analysis. All analyses were done on a NovoCyte Quanteon flow cytometer.
For Incucyte cell death assays, cells were electroporated to knock out the indicated genes and cultured in 96-well plates at 1,000 cells per 100 μl per well for 24 h prior to addition of either Cytotox Red or Annexin V Green (Sartorius) dye. The Cytotox Red dye was diluted 1:2,000 in RPMI to generate a 500 nM solution; 100 μl were added to each well for a final concentration of 250 nM Cytotox Red per well. The Annexin V Green dye was solubilized in 100 μl PBS and diluted 1:100 in RPMI before adding 100 μl to each well to generate a final dilution of 1:200. Cell growth and fluorescence were monitored over time using the Sartorius Adherent Cell-by-Cell Analysis Module.
Transcriptomic analysis
Total RNA was collected from cell pellets using RNAeasy columns (Qiagen). Libraries were prepped using the NEB Ultra II RNA library prep kit and sequenced on NextSeq HighOutput 300-Cycle flow cell (2× 100bp). Data were analysed as follows. Reads were aligned using Illumina DRAGEN v4.0. Counts were calculated using featureCounts from subread package and subsequently adjusted with TMM normalization and limma-voom transformation. Differential expression analysis was performed using limma. Gene set enrichment analyses were performed using the fgsea package.
Molecular modelling of PELO–HBS1L complex
A model of the PELO–HBS1L interaction was generated by first extracting the 3D coordinates for gene PELO (chain HC, named by authors as II]) and for gene HBS1L (chain IC, named by authors as JJ) from the experimental ribosomal complex published under Protein Data Bank ID 5LZZ (electron microscopy). The electron microscopy structure displays every residue in the full-length PELO protein (1–385), but the first 259 residues of HBS1L are not resolved, displaying only residues 260–684 of the 684-amino acid HBS1L protein. Coordinates were then imported into the visualization program PyMol (version 1.7.6.6, Schrodinger). Protein backbone ribbon styling, background settings, key residue highlights (with stick) and sectional colouring were applied.
HBS1L immunoprecipitation
Cells were washed on ice in cold PBS and collected in cold IP lysis buffer (Pierce) supplemented with cOmplete mini EDTA free protease inhibitor and phosSTOP mini phosphatase inhibitor (Roche). Following lysis at 4 °C under constant agitation and centrifugation at 4 °C and 12,000 r.p.m., lysate concentration was quantified using Pierce BCA Protein Assay reagent (ThermoFisher Scientific). Lysate (1–2 mg) was incubated overnight under constant agitation at 4 °C with either HiBIT monoclonal (Promega) or mouse IgG antibody (Sigma-Aldrich); remaining lysate was reserved for input samples. The next morning, lysates were incubated with Dynabeads protein G (ThermoFisher Scientific) for 1 h under constant agitation at 4 °C and washed 3 times with IP lysis buffer. To elute, beads were incubated at 95 °C for 5 min in 1× Protein Simple master mix consisting of dithiothreitol, sample buffer and florescent master mix. Simple Western was performed according to manufacturer’s protocol, and results were analysed using Compass software (version 6.2.0).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.