DNA
Oligonucleotides were purchased from Integrated DNA Technologies with standard purification and desalting. See Supplementary Data for all oligonucleotides and DNA constructs.
Media
Unless otherwise stated, all cultures were grown in LB-Lennox medium (10 g l−1 bacto tryptone, 5 g l−1 sodium chloride, 5 g l−1 yeast extract). LB agar plates were composed of LB plus 15 g l−1 bacto agar. M9 minimal medium (12.8 g l−1 Na2HPO4, 3 g l−1 KH2PO4, 1 g l−1 NH4Cl, 0.5 g l−1 NaCl, 3 mg l−1 CaCl2) was adjusted to pH 7.5 with 10 M NaOH. For phage experiments, Tryptone-KCl (TK) liquid medium comprised of 10 g l−1 Tryptone, 5 g l−1 KCl and 0.5 ml l−1 of 1 M CaCl2. TK sloppy agar contained 10 g l−1 Tryptone, 5 g l−1 NaCl and 7 g l−1 agar. TK bottom agar contained 10 g l−1 Tryptone, 2.5 g l−1 NaCl, 2.5 g l−1 KCl, 0.5 ml l−1 of 1M CaCl2 and 10 g l−1 agar. Super optimal broth with catabolite repression (SOC) liquid medium contained (20 g l−1 tryptone, 5 g l−1 yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4 and 20 mM glucose). For protein yield quantification, terrific broth medium (TB) contained 11.8 g l−1 tryptone, 23.6 g l−1 yeast extract, 9.4 g l−1 K2HPO4, 2.2 g l−1 KH2PO4 and 0.8% v/v glycerol.
Selective agents
ColE1 was expressed in strain JC411 and purified as previously described61. All other selective agents were purchased commercially: carbenicillin (50 µg ml−1), chloramphenicol (40 µg ml−1), gentamycin (10 µg ml−1), kanamycin (30 µg ml−1), sodium dodecyl sulfate (SDS) (0.005% w/v), spectinomycin (190 µg ml−1), tetracycline (15 µg ml−1), hygromycin (150 µg ml−1), zeocin (10 µg ml−1) and colicin E1 (ColE1; ~10 µg ml−1).
Promoter inducers
Plasmid-induced pORTMAGE62 and Recombineering63: l-arabinose (0.2% w/v, pBAD promoter), rhamnose (0.15% w/v, rhaB promoter). Plasmid maintenance was carried out with tetracycline (5–10 ng ml−1, tetA selectable marker).
OTS expression for nsAA incorporation assays: l-arabinose (0.05% w/v, pBAD promoter) and anhydrotetracycline (100 ng ml−1, pL tetO promoter).
Non-standard amino acids
BocK was purchased from Chem-Impex (00363) and dissolved in LB or TB to a final concentration of 10 mM. pAcF was purchased from Chem-Impex (24756), dissolved in sterile water to a concentration of 50 mM and used at a final concentration of 1 mM.
Strains
All strains were built on rEc∆1.∆A-C321.ΔA4 (MG1655 dnaG.Q576A.exoX.K28TAA.ΔtolC.ΔprfA.ΔmutS::zeo.Δ (ybhB-bioAB)::[λcI857 N(cro-ea59)::tetR-bla].12B.tolQRA); with all instances of TAG codons converted to TAA and 1,195 naturally occurring TGA codons.
Selectable marker preparation
Selectable markers were amplified by PCR (40 µl per reaction) performed using Kapa HiFi HotStart ReadyMix according to the manufacturer’s protocols with annealing at 61 °C. Primers were designed via Benchling primer creation software, confirmed with Kun’s oligonucleotide Tm calculator (http://arep.med.harvard.edu/kzhang/cgi-bin/myOligoTm.cgi). PCR products were purified using a Qiagen PCR purification kit, eluted in 30 µl dH2O and quantified either using an Eppendorf BioPhotometer plus or NanoDrop ND1000 spectrophotometer. For analysis, amplicons were run on a 1.5% agarose gel stained with ethidium bromide to confirm the expected band sizes.
TGA-to-TAA recoding
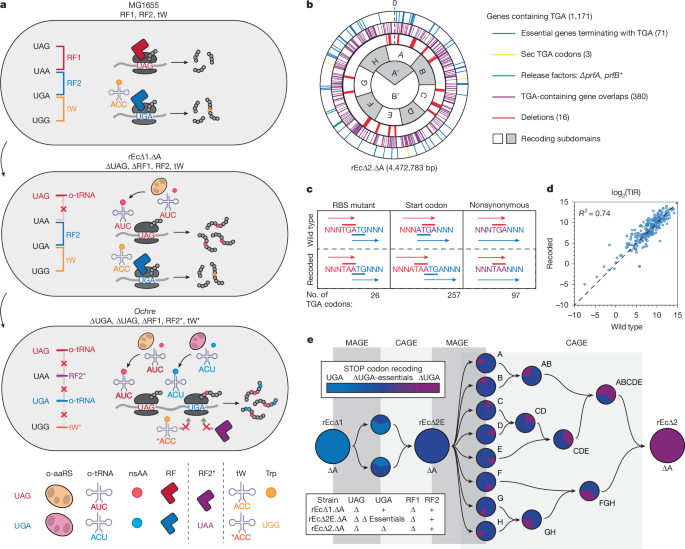
The starting strain was a standard rEc∆1.∆A (C321.∆A) with a zeocin marker cassette replacing a chloramphenicol marker at the mutS locus (Supplementary Table 10). The ybgC-tolQRA locus was duplicated between the ycgV and ychF genes as previously described64 to increase the fidelity of ColE1 selections. The previously reported mutations to dnaG and exoX to improve MAGE efficiency were also present in the starting strain65. All genes containing TGA codons were identified from WGS (Supplementary Table 1; see Methods section ‘WGS and analysis’ below). Eliminating TGA codons required a combined strategy of MAGE-mediated conversions of TGA to TAA and non-essential gene deletions (Supplementary Table 7). The oligonucleotides were designed as previously described30, ordered from IDT and grouped into pools up to 11 oligonucleotides per pool at 10–12 µM total DNA per pool, regardless of number of oligonucleotides (N) (Supplementary Table 1). These pools were used for N + 1 rounds of MAGE before cultures were plated. MAGE was performed as previously described30. Forty-seven colonies were picked and screened using Multiplex allele-specific colony PCR (MASC-PCR), as previously described15. Simultaneously, picked colonies were grown to confluence, diluted 1:15 into 150 µl of M9 minimal medium, then inoculated 1:100 into 150 µl LB and M9 to assess growth phenotypes (MaxOD, doubling time and lag time) (see ‘Fitness analysis’). The colony containing the largest number of lowest frequency conversions and minimal deviations in growth fitness was chosen for subsequent rounds of MAGE.
Native RBS predictions
Predicted RBS translation rates of genes overlapped by TGA stop codons before and after recoding to TAA were calculated in alignment with previous protocols36.
MAGE and λ-Red-mediated recombination
MAGE, pORTMAGE and λ-Red-mediated recombination were performed as previously described15,62. Cells were transferred to 0.1 cm cuvettes, electroporated (BioRad GenePulser, 1.78 kV, 200 Ω, 25 µF) and immediately resuspended into 3 ml LB (MAGE) or 1 ml SOC medium (dsDNA), grown at 34 °C, 225 rpm. For tolC and galK negative selections, cultures were recovered for at least 7 h to allow complete protein turnover before exposure to ColE1 and 2-deoxygalactose, respectively. Once all strains were conjugated into a final strain (see ‘CAGE assembly’), genomically integrated λ-Red was displaced by tolC negative selection. To convert background mutations and remaining TGA codons we employed a plasmid-based single stranded DNA recombineering approach. l-arabinose-inducible λ-Red-derived recombineering machinery was expressed on a temperature-curable plasmid. For double stranded DNA (dsDNA) recombination, rhamnose-inducible episomal λ-Red-derived recombineering machinery was also expressed on a temperature-curable plasmid containing l-arabinose-inducible ccdA antitoxin for negative selections as previous described63 for both marker removal and release factor placement/displacement. Both λ-Red plasmids contained tetA selectable markers. To cure plasmids, strains were incubated with λ-Red induction at 37 °C overnight in 3 ml LB, plated onto solid medium, incubated at 42 °C for 2–4 h, then incubated at 37 °C until colonies were visible. Colonies were picked into 150 µl LB, incubated for 3 h, then transferred 10 µl into LB with tetracycline to identify colonies lacking plasmids.
Genotyping
MASC-PCR was used to simultaneously detect up to 11 TGA-to-TAA conversions or background mutation reversions, as previously described with TAG conversions15. All primers are designed with a 61 °C annealing temperature, according to Kun’s oligonucleotide Tm calculator, while avoiding 3′ end binding in secondary structure, according to Benchling’s primer secondary structure prediction function. Each reaction consisted of KAPA 2 G Fast Multiplex ReadyMix (Kapa Biosystems, KK5802), 2 µl of template DNA and 0.2 µM of each primer for each 10 µl reaction. MASC-PCR results were run on 2.2% agarose gels with ethidium bromide staining. After λ-Red-mediated recombination or conjugation, colony PCR was used to confirm the presence and absence of selectable markers at desired positions. Colony PCR (10 µl per reaction; annealing at 61 °C (Kun’s oligonucleotide Tm calculator)) was performed using Kapa 2G Fast HotStart ReadyMix following manufacturer’s protocols. Results were analysed on a 1.5% agarose gel stained with ethidium bromide. Sanger sequencing was performed by Genewiz.
CAGE assembly
Conjugative assembly genome engineering (CAGE) was performed using the protocol as previously described15. Deviating from previous protocols, the donor strain had two positive markers—a spectinomycin marker and a gentamycin marker, each flanking the recoded region, in addition to the kanamycin resistance origin of transfer (kanR-oriT) cassette set ~3–5 kb upstream (Extended Data Fig. 2 and Supplementary Table 10). The donor strain also contained a modified RK24 plasmid in which all genes that end in a TAG codon were recoded to TAA10. Cell spots were rinsed twice with 500 µl LB and collected to give a 1 ml 10−2 dilution of conjugated cells, followed by an additional 10-fold dilution in a new tube. 50 µl of 10−2 and 10−3 dilutions were each plated onto LB agar plate with appropriate antibiotics to select for both the recipient background selectable markers and the donor region selectable markers. Forty-seven candidate colonies were grown in a 96-well format and screened for desired genotypes via PCR (to confirm presence and absence of selectable markers) and MASC-PCR (to confirm the presence of interspersed desired codon replacements). For large genomic transfers (>1 Mb), final strains were sequence verified through WGS (see ‘Genotyping’). The resulting strain contained the three selectable markers, all of which were subsequently deleted via dsDNA λ-Red tolC-mediated selection–counterselection (via SDS or ColE1 selection, respectively), or maintained for the next conjugation64.
Genomic deletions
To reduce the overall number of genes requiring ∆TGA recoding via MAGE, 16 multigenic regions containing a total of 229 genes (3 later categorized as pseudogenes) were identified for deletion based on previous work by the Blattner laboratory29 (Extended Data Fig. 1). Each targeted deletion site (up to ~34 kb in size) was displaced via tolC selectable marker displacement and counterselection for subsequent marker removal in accordance with previous protocols64. ColE1 selections on solid medium were performed as previously described15,64. For pre-selection, 5 µl of recovered cultures were inoculated into 150 µl LB with either carbenicillin (control) or ColE1 with vancomycin (64 µg ml−1), in triplicate, in a 96-well plate and incubated at 34 °C for 16 h to monitor growth along the progenitor strain with tolC. Strains exhibiting growth in both carbenicillin and ColE1 with vancomycin were considered positive for tolC deletion and subsequently plated onto LB agar plates with ColE1 for monoclonal colony selection and PCR screened to confirm the loss of tolC. Subsequent MASC analyses allowed for selection of strains with both deletion events and TGA conversions to reduce the total number of MAGE cycles. After every tolC placement and gene displacement, strain growth curves were analysed to assess gene deletion impact on strain phenotype (see ‘Fitness analysis’). Major growth impairments were assessed. Additional major deletions (>50 bp) resulted either from mutagenesis of highly repetitive noncoding genes, active transposable elements or scarring from conjugations.
WGS and analysis
Whole genomes were isolated using the Qiagen DNeasy Blood and Tissue isolation kit. For high fidelity genome analysis performed after conjugations, raw reads were acquired via short-read 150-bp (50× coverage) paired-end Illumina sequencing data were collected with Hiseq 4000 with libraries prepared by the Yale Center for Genome Analysis. Long-read (<25 kb, 50× coverage) data were prepared by Pacific Biosciences Single Molecule Real-Time (SMRT) Analysis. For rapid whole-genome analysis, raw reads were acquired via Plasmidsaurus Oxford nanopore standard bacterial WGS (30× coverage) via bacterial pellet submissions suspended in DNA/RNA Shield. To perform analyses on raw reads, latest versions of breseq 0.38.1 computation pipeline were employed for aligning sequence reads to rEc1.∆A (C321.∆A) reference genome, run using Ubuntu LTS on Windows Subsystem for Linux, in accordance with https://barricklab.org37. Summary .html outputs provided comprehensive lists of mismatches, indels and missing or novel junctions to monitor TGA conversion progress, deletions and background mutagenesis. Mutations were then organized by type in Excel (see Supplementary Tables).
AlphaFold structure of RF2.B3
The 3D structure of RF2.B3 in Fig. 2b was acquired through Benchling AlphaFold48 online prediction from amino acid sequence submission (Supplementary Table 17) and spatially oriented for view of primary reaction sites.
Liquid selection complementation
In triplicate, strains harbouring prfB variant expression plasmids (as described in Extended Data Fig. 4 and Supplementary Discussion 1) were electroporated with a kanamycin resistance (kanR) dsDNA cassette for displacement of genomic prfB and recovered in varying concentrations of vanillic acid inducer for 3 h at 37 °C to allow episomal expression of prfB variants. Strains were then diluted 1:50 into selective LB medium 25 µg ml−1 kanamycin and incubated with shaking at 37 °C, monitoring absorbance at 600 nm in a BioTek Synergy H1 plate reader (Agilent). After 36 h of growth, OD600 for each strain was reported as viability after complementation. Individual colonies were isolated by plating knockout strains on selective medium after recovery and PCR screened to confirm kanR displacements of genomic prfB. Colonies that tested positive with PCR screen were confirmed by WGS (Plasmidsaurus) to not harbour genomic prfB or secondary mutations, and whole-plasmid sequencing (Plasmidsaurus) of their prfB-containing plasmid to confirm variant.
Readthrough fluorescence assay
We used a dual fluorescence mCherry–YFP reporter to test stop codon readthrough as previously reported66. This reporter was constitutively expressed from a low copy backbone (p15A) to maximize dynamic range for detecting post-transcriptional fluctuations. Each evaluated strain was transformed with four variants of the reporter (three stop codons and a sense codon positive control GCG) and plated on selective medium for overnight growth. Colonies were picked in triplicate into 150 µl cultures of LB with appropriate selection in 96-well plates, grown with shaking at 225 rpm in 37 °C for 18 h after which timepoints measurements were taken for mCherry (excitation: 585 nm, emission: 635 nm, gain: 100), YFP (excitation: 500 nm, emission: 541 nm, gain: 80) and OD600 (absorbance: 600 nm) in a BioTek Synergy H1 plate reader (Agilent). To assess cognate amber or opal suppression, a plasmid bearing supD52 with CUA or UCA anticodon was co-transformed with the dual fluorescence reporter plasmid. Fluorescence values of all strain and plasmid conditions were normalized to OD600 before dividing by the positive control described above to arrive at fractional mCherry and YFP signals reported.
Bacteriophage assays
For all phage experiments, growth was carried out in TK at 37 °C; infection and propagation occurred in TK sloppy agar poured onto solid TK bottom agar incubated at 37 °C. TK sloppy agar mixes were maintained at 45 °C before use.
Phage propagation
For propagation, E. coli MG1655 was grown to mid-log phase in 3 ml of LB. Two-hundred microlitres of bacteria was added to 3 ml TK sloppy agar. Immediately following, 50 µl of phage (10 to 100-fold dilutions) was added directly from refrigerated stock into the sloppy-bacterial culture. Three millilitres of sloppy bacteria phage culture was poured onto solid TK bottom agar plates, dispersed evenly, then left at room temperature to solidify. Plates were then incubated at 37 °C for ~16 h to permit lysis to proceed to completion. The entire sloppy agar was collected and centrifuged (12,000g, 2 min) and 3 ml of supernatant was filtered with 0.22-µm filter column to remove bacteria.
Phage titration
Bacteria strains were grown overnight at 37 °C until OD600 reached 2–3. Twenty microlitres of bacteria was added to 3 ml sloppy agar. Three millilitres of sloppy bacteria culture was poured onto solid TK bottom agar plates. After incubated at room temperature for 15 min, 3 µl of the phage dilutions (101– to 108-fold dilutions) were dropped on the surface of the solidified sloppy agar. Once the drops dried, plates were incubated overnight at 37 °C. Visible plaques were counted for the individual drops. Titres (in PFU per ml) were calculated according to \({\rm{PFU}}\,{\rm{per}}\,{\rm{ml}}=N\times \frac{1}{{\rm{DF}}}\times \frac{1}{V}\), where N is the number of plaques, DF is the phage dilution factor and V is the volume of phage dilution pipetted on the plate.
Fitness analysis
Kinetic growth curves
Kinetic growth (OD600) curves were obtained via monitoring strain growth within a BioTek Synergy HT plate reader. Each strain was grown in triplicate within 96-well plates, 150 µl LB and 150 µl M9, within 96-well flat-bottom plates, incubated at 34 °C for intermediate λ-Red+ strains and 37 °C for final strains, 225 rpm, for 16–36 h and absorbance at 600 nm was read at 10-min intervals. In preparation, strains were grown to confluence in LB, diluted 1:15 into 150 µl of M9 minimal medium and inoculated 1:100 into 150 µl LB and M9. Auxotrophic strains revealed no growth in M9.
OD600 calibration
The absorbance obtained by the BioTek Synergy HT plate reader was recalibrated to OD600 (absorbance at 600 nm through 1 cm pathway) using a standard curve \(y=2.746x+1.878{x}^{2}\). The OD600 of an overnight LB culture of MG1655 was measured with a Biochrom Libra S4 Spectrophotometer at 600 nm wavelength in a semi-micro cuvette (1 cm pathway) after 1:10 dilution of culture into 1 ml LB. To generate the calibration curve, a series of cultures with OD600 ranging from 0 to 6 were prepared by diluting in LB medium. These cultures were then measured by the BioTek Synergy HT plate reader, with the same settings as growth cultures. The average values were then fitted to a polynomial standard curve for recalibration. The effects of medium evaporation in plate wells are not considered.
Doubling time and MaxOD calculation
The recalibrated growth curve was used to calculate the doubling time and MaxOD. Linear fitting of log2 OD600 was performed using a sliding-window method67, where the window size is 50 min for rich medium and 100 min for M9 minimal medium in the early log phase, respectively. The doubling time was calculated as the reciprocal of the slope. MaxOD was obtained within the 36 h growth period. Individual biological sample n ≥ 4.
nsAA incorporation assays
Plasmid construction
Gene fragments for aaRS and tRNA were synthesized by Twist Bioscience and cloned into expression vectors by Golden Gate assembly. Plasmids were sequence verified by whole-plasmid sequencing (Plasmidsaurus or Quintarabio). All cloning was made in Mach1 (Thermo Fisher, C862003).
Incorporation of BocK and pAcF into proteins
Recoded strains were transformed with OTS-reporter plasmids by standard electroporation protocols. Electroporated strains were recovered in 2 ml LB or SOC for at least 2 h before plating onto LB agar plates with kanamycin (50 µg ml−1) and incubated at 37 °C overnight. Three single colonies from each plate were picked and grown in 800 µl LB supplemented with kanamycin (50 µg ml−1) in a 96 deep-well plate sealed with a Breathe-Easy film (Sigma-Aldrich) and incubated at 37 °C with shaking at 220 rpm for 20–24 h. For nsAA incorporation at UAG and/or UGA, after overnight growth the cultures were back-diluted 1:50 onto a clear-bottom black 96-well plate (Costar) in a total of 150 µl of LB supplemented with kanamycin (50 µg ml−1), aTc (100 ng ml−1), l-arabinose (0.05% w/v), 1 mM pAcF and/or 10 mM BocK. Cell growth (absorbance at OD600) and GFP fluorescence (excitation 485 nm, emission 525, gain 70, bottom measurement) were measured in a BioTek Synergy H1 plate reader (Agilent) for 24 h at 10 min intervals with linear shaking. Data were analysed with a custom Python script. All reported GFP fluorescence was normalized to ELP–3×TAC–GFP control fluorescence within each corresponding condition.
Expression of ELP–3×(TGA-TAG)–GFP with nsAAs for mass spectrometry
Single colonies of rEc∆2.∆A.B3 and rEc∆2.∆A.B3.tW* (Ochre) containing the dual-OTS dual reporter plasmid were inoculated in 2 ml LB with 50 µg ml−1 kanamycin in a 14 ml falcon tube overnight at 37 °C with shaking at 220 rpm. After overnight growth the cultures were diluted 1:100 in 25 ml LB-kanamycin and grown until OD600 at 0.6, where the cultures were supplemented with 1 mM pAcF and 10 mM BocK and induced with 100 ng ml−1 aTc and 0.05% w/v l-arabinose. The cultures were grown at 37 °C overnight, after which the cells were collected by centrifugation at 3,200g for 20 min in a 50 ml centrifuge tube and stored at −80 °C until protein purification.
Protein mass spectrometry
ELP–GFP reporter protein purification
Frozen E. coli cell pellets were thawed on ice and pellets were lysed by sonication with lysis buffer consisting of 50 mM Tris-HCl (pH 7.4, 23 °C), 500 mM NaCl, 0.5 mM EGTA, 1 mM DTT, 10% glycerol, 50 mM NaF and 1 mM Na3O4V. The extract was clarified with two rounds of centrifugation performed for 20 min at 4 °C and 14,000g. Cell-free extracts were applied to Ni-NTA metal affinity resin and purified according to the manufacturer’s instructions. Wash buffers contained 50 mM Tris pH 7.5, 500 mM NaCl, 0.5 mM EGTA, 1 mM DTT, 50 mM NaF, 1 mM Na3VO4 and 10 mM imidazole. Proteins were eluted with a wash buffer containing 250 mM imidazole. Eluted protein was subjected to 4 rounds of buffer exchange (20 mM Tris pH 8.0 and 100 mM NaCl) and concentrated using a 30 kDa molecular weight cut-off spin filter (Amicon).
Protein digestion and mass spectrometry
Affinity purified, buffer exchanged ELP–GFP reporter protein, or whole cell lysates, were digested and analysed by mass spectrometry as described previously with some modifications55,68,69. ELP–GFP reporter protein (5–10 μg) was diluted with water and 20% SDS for a final volume of 115 μl and final concentration of 1% SDS. Samples were denatured for 15 min at 55 °C in a heat block. Reduction and alkylation of cysteines was performed with TCEP and 2-chloroacetamide (CAM) using a final TCEP and CAM concentration of 10 mM and 44 mM respectively. The reduction-alkylation reaction proceeded for 20 min at 55 °C. 6 μl of 50 mg ml−1 SP3 beads (Speed Bead, Cytiva) pre-washed and resuspended with water were added to samples for a final working volume of 134 μl. Binding of protein to the beads was induced by adding 150 μl of 100% ethanol. The binding mixture was incubated in a ThermoMixer Eppendorf at 24 °C for 10 min at 1,400 rpm. After binding, the beads were magnetized on a magnetic rack and supernatants were removed. This was followed by three rounds of bead washes with 500 μl of 80% ethanol per wash. All traces of 80% ethanol were removed after the last wash. Beads in each sample were resuspended with 50 μl of digestion solution containing 0.4 μg of sequencing grade trypsin (Promega) in 50 mM triethylammonium bicarbonate (TEAB) buffer (Sigma). Digests were incubated for 16 h at 37 °C in a ThermoMixer at 1,400 rpm. Beads were magnetized and 50 μl supernatants were moved to fresh tubes. Beads were resuspended in 50 μl of 50 mM TEAB buffer and incubated for 5 min at 37 °C in a ThermoMixer at 1,400 rpm to maximize peptide recovery. Beads were magnetized again and the two 50 μl supernatants were combined. Peptides were dried in a vacuum centrifuge at room temperature. Dried peptides were reconstituted in 2/98 acetonitrile/water with 0.1% formic acid and analysed by LC–MS/MS. LC–MS/MS was performed using a Vanquish Neo UHPLC system (Thermo) and an Orbitrap Eclipse Tribrid Mass Spectrometer (Thermo). The analytical column employed was a 75 μm inner diameter, fused silica capillary tube (Molex) packed in-house to a length of 15 cm with 1.9 μm ReproSil-Pur 120 Å C18-AQ (Dr. Maisch) using methanol as the packing solvent. Column was attached to a PepSep Spray Adapter with a fused silica emitter (Bruker). Peptide separation was achieved using mixtures of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) with a 41-min gradient; 0/5, 30/30, 39/45, 40/55, 41/100 (time (min)/B (%), linear ramping between steps). The gradient was performed with a flowrate of 300 nl min−1. At least one blank injection (5 μl 2% B) was performed between samples to eliminate peptide carryover on the analytical column. One-hundred nanomoles of trypsin-digested BSA and 100 ng of trypsin-digested HeLa protein standard were run periodically between samples as quality control standards. The mass spectrometer was operated with the following parameters: (MS1) 60,000 orbitrap resolution, 250% normalized AGC target, 50 ms maximum injection time, 300–1,400 m/z scan range; (data dependent-MS2) Ion Trap detector, 200% normalized AGC target, 13 ms maximum injection time, top 10 mode, 1.2 m/z isolation window, 30% normalized HCD collision energy, 40 s dynamic exclusion. Data were searched using MaxQuant version 1.6.10.43 with deamidation (NQ), oxidation (M) and phospho (STY) as variable modifications and carbamidomethyl (C) as a fixed modification with up to 3 missed cleavages, 5 amino acids minimum length and 1% false discovery rate against a modified Uniprot E. coli database containing custom MS-READ reporter proteins. MS-READ search results were analysed using MaxQuant and Perseus version 1.6.2.2
Protein yield quantification
Protein expression
Precultures were inoculated from fresh colonies picked from selective LB agar plates (LB plates + antibiotics at 37 °C for 24 h) into 2 ml TB with 50 µg ml−1 kanamycin and grown overnight at 37 °C with shaking at 220 rpm. Overnight precultures were diluted to OD600 of 0.1 in 20 ml TB with 50 µg ml−1 kanamycin and grown until OD600 = 0.6–0.8. pAcF was added to a final concentration of 1 mM. Reporter and OTS expression was induced with 100 ng ml−1 aTc and 0.05% arabinose. Cells were collected 8 h post-induction by centrifugation at 4,000g and the supernatant was removed. Cell pellets were resuspended in 1 ml wash buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.5), transferred to 1.7 ml tubes and centrifuged at 17,000g. The wash buffer was then removed and the cell pellet stored at −80 °C until further use.
Lysis and extraction
Pellets were resuspended in sufficient volume of wash buffer to normalize OD600 to 10. 1 ml of cells were lysed using Sigma-Aldrich 1× BugBuster (from 10× stock) and centrifuged at 16,000g to isolate soluble fraction.
Quantifying GFP concentration
GFP quantification methods were adapted from previous work70. An ELP–sfGFP standard with an N-Terminal 6×His tag (containing no in-frame stop codon and cloned in a pET28a vector) was expressed in BL21. The ELP–sfGFP standard was purified using a Ni-NTA column and the protein was quantified using a Pierce BCA (Bicinchoninic Acid) Protein Assay kit (Thermo Fisher) using established methods68. A GFP standard curve was produced by spiking the purified GFP standard into lysate prepared from Ochre that contained no GFP. GFP was spiked in at 28, 18.6, 12.5, 8.3, 5.5, 3.7, 2.5, 1.6, 1.1, 0.7, 0.5 and 0.3 µg ml−1 to generate triplicate, linear standard GFP curves. Lysates of different GFP expressing strains were prepared in a similar fashion and dilutions were fit to the linear range of the standard curve. Fluorescence of clarified lysates was measured in an H1M BioTek plate reader (excitation: 485 nm, emission :510 nm, gain: 60) alongside the GFP standard. Fluorescence values were converted to µg ml−1 and used to calculate total yields of expressed protein.
Statistical analysis
Statistical significance was typically generated from two-tailed unpaired t-tests. P values for mCherry–YFP readthrough assays (Fig. 2e,f and Extended Data Fig. 5e) were calculated using Welch’s unpaired t-test by comparison to rEc∆1.∆A within each paired codon and suppressor condition. Error bars display s.e.m., n = 3. P values for phage infection assays (Fig. 3) and tRNATrp UGA suppression assays (Fig. 4c,d) were derived from unpaired t-tests by comparison to E. coli MG1655 within each graph. Error bars display 95% confidence interval, n = 3. P values for tRNATrp UGA suppression assays (Fig. 4c) were derived from unpaired t-tests in relation to no Mj-TyrOTS plasmid within each strain. Error bars display 95% confidence interval, n = 3. P values for strain doubling times and maximum OD600 were calculated using Mann–Whitney U-tests comparing strain values with those of rEc∆1.∆A within each graph. Error bars display 95% confidence interval, n = 7 or 11 replicates (Fig. 5), n = 8 (Extended Data Fig. 3, LB) or n = 4 (Extended Data Fig. 3, 2×YT and TB) replicates. P values in ELP–GFP fluorescence assays assessing nsAA incorporation efficiencies (Fig. 6b–d and Extended Data Fig. 9c–e) were derived from unpaired t-tests comparing data to rEc∆1.∆A within each nsAA condition. Error bars display 95% confidence interval, n = 3. P values for RF2 complementation fluorescent protein production (Extended Data Fig. 4f and Supplementary Fig. 3a) compare fluorescence of GFP expressed from RF2 B2(P205) complemented strains to fluorescence of B1(wild type (S205)) strains with a similar genotype and vanillic acid concentration using unpaired t-tests (n = 3). Error bars display s.e.m. P values comparing RF2.B1 and B2 TGA readthrough (Supplementary Fig. 3b) for various vanillic acid concentrations compared to 0.1 µM vanillic acid within each strain release factor variant were calculated by Welch’s t-tests (n = 3). Error bars display s.e.m. Calculations and graphs were generated using Graph Pad Prism (v.10.1.0). Figures were prepared using Adobe Illustrator 2023 (v.28.2). All measurements were taken from distinct samples except when measured repeatedly over time to collect time courses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.