Cell lines and cell culture
HEK 293T cells (ATCC) and TZM-bl cells (NIH HIV Reagent Program) were maintained in Dulbecco’s modified Eagle medium (Thermo Fisher Scientific) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco) and 1% penicillin–streptomycin at 37 °C and 5% CO2. Expi293F cells were maintained in Expi293 Expression Medium (Thermo Fisher Scientific) in Erlenmeyer flasks at 37 °C, 5% CO2, 125 rpm. Cell lines were authenticated by the organizations from which they were obtained: 293T cells by STR profiling, Expi293F cells by STR profiling and assessment of cell morphology and growth kinetics, and TZM-bl cells by assessment of cell morphology and functional testing in neutralization assays. Cell lines were not routinely tested for mycoplasma contamination.
Antibody expression and purification
The ITS01, ITS06.02, ITS102.03 and ITS103.01 (both LS and non-LS versions for the latter two) mAbs were produced by cotransfection of plasmids encoding heavy and light chains for a given antibody at a 1:1 mass ratio into Expi293F cells (Thermo Fisher Scientific) using an ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were cultured at 37 °C for 6 days following transfection and then moved to 32 °C for the final day 7 of culture. Culture supernatant was clarified by low-speed centrifugation and 0.22-μm filtration. Antibodies were purified using rProtein A Fast Flow resin (Cytiva), buffer exchanged into phosphate-buffered saline (PBS) and sterile filtered. Antibody preparations were stored at 4 °C or frozen at −80 °C.
Virus stock production
SIVsmE660.FL14-IAKN virus (a molecular tier 2 clone of SIVsmE660) was initially prepared in HEK 293T cells transfected with SIVsmE660-encoded plasmid pUC19-E660-FL14 using FuGene6 (Promega). Two days posttransfection, the cell supernatant was collected (transfection stock). Rhesus macaque peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using SepMate tubes (StemCell Technologies) with Ficoll (Cytiva) density gradient centrifugation. CD4+ T cells were enriched by positive selection (CD4 MicroBeads, NHP, Miltenyi Biotec). Rhesus CD4+ T cells were activated by incubation with α-CD2/α-CD3/α-CD28 microbeads (T Cell Activation/Expansion Kit, NHP, Miltenyi Biotec) in 30 U ml−1 IL-2 in RPMI medium containing 15% (v/v) heat-inactivated fetal bovine serum and 1% penicillin–streptomycin (RF15-IL2) for 4 days. The rhesus macaque PBMCs were separately isolated and activated from 3–5 animals. The activated CD4+ T cells were harvested, pooled and infected with the transfection stock at a multiplicity of infection of 0.1–0.01. Cells were incubated at 37 °C and 5% CO2 for 4 h with gentle mixing every 30 min and washed with R15-IL2 three times. Then, the infected cells were transferred to tissue-culture-treated flasks and cultured for 15 days. During the 15 days, 200 µl of culture supernatant was collected and frozen every 3 days beginning at day 0 for p27 ELISA (Advanced Bioscience Laboratories). About 75% of culture supernatant was collected and frozen at days 6, 9, 12 and 15, and the same volume of warm R15-IL2 was added to the flask. Supernatants with relatively high p27 values were thawed, aliquoted and stored at −80 °C (infection stock). Individual infectious stocks of the molecularly tagged (barcoded) SIVmac239 viruses A to I were generated as described previously30.
Animal studies
All experiments were carried out in compliance with National Institutes of Health regulations and with approval from the Animal Care and Use Committee of the Vaccine Research Center and Bioqual, Inc., where NHPs were housed for the duration of the studies. Animals were housed and cared for in accordance with local, state, federal and institutional policies in facilities accredited by AAALAC International under standards established in the Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals. In accordance with the institutional policies of both institutions, all compatible NHPs were always pair-housed; single housing is only permissible when scientifically justified or for veterinary medical reasons, and for the shortest duration possible. NHPs were housed in appropriately sized caging according to the Guide for the Care and Use of Laboratory Animals40 and provided with a variety of enrichment toys, treats, fresh produce and foraging devices. Water was offered ad libitum, and animals were fed primate biscuits (Monkey Diet, 5038, Lab Diet) twice daily. As standard practice, animal holding rooms were maintained on a 12-h light–dark cycle, with a room temperature of 16–21 °C and relative humidity of 30–70%. The investigators were not blinded to the animal treatment allocations.
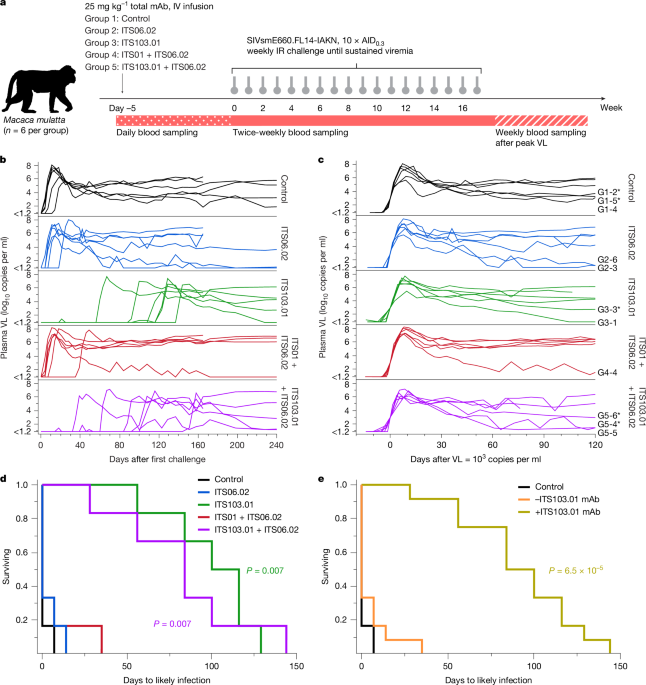
In the partially and fully neutralizing mAb infusion and challenge study, 30 Indian-origin rhesus macaques (Macaca mulatta), aged 2–5 years, were stratified by sex, age, weight and TRIM5 genotype into five groups of six animals (Supplementary Table 5). Distribution of TRIM5 genotypes was required as SIVsmE660 acquisition is affected by certain alleles41,42. Sample size was chosen to yield at least 85% power to detect a difference in time to infection (this also applied to the barcoded virus study described below). Animals were intravenously infused with ITS06.02 (group 2), ITS103.01 (group 3), ITS01 and ITS06.02 (group 4), or ITS103.01 and ITS06.02 (group 5). All animals received a total of 25 mg kg−1 of mAb. For groups 4 and 5, the mAbs were given at a mass ratio of 1:1. The mAbs used all contained the LS mutation43. Control animals (group 1) were untreated. Five days after infusion, animals were intrarectally challenged with a limiting dose 10-fold over the animal infectious dose leading to infection of 30% of control animals (10 × AID0.3) of SIVsmE660.FL14-IAKN infection stock. For each animal, these challenges were repeated weekly for a maximum of 17 weeks until sustained viremia occurred. Blood samples were collected throughout the study.
The barcoded virus challenge study used 16 Indian-origin rhesus macaques (M. mulatta), aged 2 years, which were stratified by sex, age and weight into one group of four animals (control, group 1) and two groups of six animals (mAb treatment, groups 2 and 3) (Supplementary Table 6). mAb-treatment animals were intravenously infused with ITS102.03 (group 2) or ITS103.01 (group 3) at 25 mg kg−1. To decrease plasma half-life to allow faster decline of mAb concentrations, mAbs that did not contain the LS mutation were used here. Control animals were not infused with mAb. Five days following mAb infusion, all groups began receiving weekly intrarectal challenges sequentially using one of eight uniquely molecularly tagged (barcoded) SIVmac239 viruses, in the order A, B, C, E, F, G, H, I. The challenge dose was equal to a dose that infects 80% of control animals (AID0.8), and challenges (up to a total of 16 by repeating the same barcodes in a second series) were continued until sustained viremia occurred in each animal. Blood samples were collected throughout the study.
Viral load measurement
Plasma SIV RNA measurements were performed essentially as described previously34. The upslope at synchronized viral load was calculated as the gradient between the plasma measurements immediately before and after viral load first passed through 1,000 copies per millilitre. The set-point viral load in each animal was calculated as the geometric mean of viral load measurements from 45 to 120 days after the viral load first reaching 1,000 copies per millilitre of plasma. Groups were statistically compared using Kruskal–Wallis and Dunn’s multiple comparisons test.
SIVmac239 barcode identification
Plasma virus barcodes were identified using either single-genome amplification as described previously30 or MiSeq sequencing as described elsewhere44. For sequence analysis, we used the Frederick National Laboratory for Cancer Research Barcode Analysis Tool v.2022 (https://frederick.cancer.gov/research/aids-and-cancer-virus-program/sections/retroviral-evolution-section or https://github.com/KeeleLab?tab=repositories), a custom algorithm written in R (v.4.3.1) for analysis of barcoded viruses.
Plasma mAb measurement
Concentrations of infused mAbs in plasma samples were determined by enzyme-linked immunosorbent assays (ELISAs) using anti-idiotypic antibodies. MaxiSorp ELISA plates (96-well, Thermo Fisher Scientific) were coated with 100 μl per well of 2 μg ml−1 anti-idiotypic antibody (17B4-IgG1, 8A4-IgG1, ITS103-id1 or ITS102-id1, as appropriate for the mAb of interest)27,28 in PBS (pH 7.4) and incubated overnight at 4 °C. Plates were blocked with 300 μl per well PBS containing 5% (w/v) skim milk powder at 37 °C for 1 h. All subsequent steps involved incubation at the same temperature and for the same time, with 100 μl loaded per well. For ITS01 and ITS06.02 quantitation assays, plasma samples were heat inactivated at 56 °C for 1 h before being diluted into PBS containing 5% (w/v) skim milk powder and 0.05% (v/v) Tween 20 (diluent buffer). For ITS102.03 and ITS103.01 quantitation assays, plasma samples were not heat inactivated before dilution into diluent buffer. Serial dilutions of the appropriate purified ITS mAb into diluent buffer were used as standards in the assay. After blocking, wells were washed five times with PBS containing 0.1% Tween 20 (and similarly washed after each subsequent step). Dilutions of plasma or mAb were then added to the plate. Bound antibody was detected using mouse anti-monkey IgG-HRP (Southern Biotech) at 1:5,000 in diluent buffer. Reactions were developed using 100 μl per well SureBlue TMB 1-Component Microwell Peroxidase Substrate, with incubation for 10 min at room temperature before addition of 100 μl per well 0.5 M sulfuric acid. Absorbance at 450 nm was measured using a SpectraMax 384 Plus Absorbance Plate Reader and analysed using GraphPad Prism v.10.0.3 (GraphPad Software). Concentrations of infused antibodies were interpolated from the standard curve using a sigmoidal four-parameter curve. Interpolated values from multiple dilutions of each plasma sample were averaged to give the final concentration.
Endogenous anti-SIV Env antibody assays
Endogenous antibody titres against Env were measured by ELISA using an anti-kappa specific antibody such that no infused mAb in the plasma sample would be detected (both ITS102.03 and ITS103.01 have lambda light chains). High-binding, half-area microplates (96-well, Corning Inc.) were coated with 50 μl per well of 1 μg ml−1 SIVmac239-foldon trimer26 in PBS, followed by incubation overnight at 4 °C. Plates were blocked with 150 μl per well PBS containing 5% (w/v) skim milk powder at 37 °C for 1 h. All subsequent steps involved incubation at the same temperature and for the same time, with 50 μl loaded per well. After blocking, wells were washed five times with PBS containing 0.1% Tween 20 (and similarly washed after each subsequent step). Plasma samples from weeks 0 to 16 after the dominant virus infection were assayed, as well as pre-immune plasma (from the day of first virus challenge). Heat-inactivated plasma samples were 1:5 serially diluted into diluent buffer starting from 1:100 (eight points total) before being added to the plate. Bound antibody was detected using goat anti-human Igκ-HRP (Millipore Sigma) at 1:2,500 in diluent buffer. Reactions were developed using 50 μl per well SureBlue TMB 1-Component Microwell Peroxidase Substrate (SeraCare), with incubation for 10 min at room temperature before addition of 50 μl per well 0.5 M sulfuric acid. Absorbance at 450 nm was measured using a SpectraMax 384 Plus Absorbance Plate Reader (Molecular Devices) and analysed using GraphPad Prism. Area under the curve values were calculated for serially diluted plasma samples. These values were normalized by subtracting each animal’s pre-immune area under the curve value.
Neutralization assays
SIV Env pseudotyped viruses were produced by cotransfection of 293T cells with plasmid DNA encoding SIV gp160 along with a luciferase reporter plasmid in the SG3 backbone containing HIV structural genes as described previously26. SIV gp160-encoding plasmids for the clone SIVmac239.cs.23 (ref. 45) were provided by D. Montefiori. Plasmids encoding SIV gp160 sequences with amino acid substitutions were synthesized by Genscript. Virus neutralization was measured using infection of TZM-bl target cells by pseudotyped virus as previously described46. Titres were calculated as either the IC50 or IC80 for mAbs that caused a 50% or 80% reduction of infection compared with virus alone using a logistic five-point model (JMP 15).
Intracellular cytokine staining
Cryopreserved PBMCs were thawed and rested overnight in a 37 °C, 5% CO2 incubator. The next morning, cells were stimulated with SIVmac239 Gag and Env peptide pools (15-mers overlapping by 11 amino acids; provided by the NIH HIV Reagent Program) at a final concentration of 2 μg ml−1 in the presence of 3 mM monensin for 6 h. Peptide pools were reconstituted in 100% dimethyl sulfoxide. Negative controls received equal concentrations of dimethyl sulfoxide instead of peptides (final concentration of 0.5%). Intracellular cytokine staining was performed as described previously47. The following mAbs were used: CD3 APC-Cy7 (clone SP34.2, BD Biosciences), CD4 PE-Cy5.5 (clone SK3, Thermo Fisher), CD8 BV570 (clone RPA-T8, BioLegend), CD45RA PE-Cy5 (clone 5H9, BD Biosciences), CCR7 BV650 (clone G043H7, BioLegend), CXCR5 PE (clone MU5UBEE, Thermo Fisher), CXCR3 BV711 (clone 1C6/CXCR3, BD Biosciences), PD-1 BUV737 (clone EH12.1, BD Biosciences), ICOS Pe-Cy7 (clone C398.4 A, BioLegend), CD69 ECD (cloneTP1.55.3, Beckman Coulter), IFN-g Ax700 (clone B27, BioLegend), IL-2 BV750 (clone MQ1-17H12, BD Biosciences), IL-4 BB700 (clone MP4-25D2, BD Biosciences), TNF-FITC (clone Mab11, BD Biosciences), IL-13 BV421 (clone JES10-5A2, BD Biosciences), IL-17 BV605 (clone BL168, BioLegend), IL-21 Ax647 (clone 3A3-N2.1, BD Biosciences) and CD154 BV785 (clone 24-31, BioLegend). LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific) was used to exclude dead cells. All antibodies were titrated to determine the optimal concentration as reported previously47. Samples were acquired on a BD FACSymphony flow cytometer and analysed using FlowJo v.10.8.2 (see Extended Data Fig. 7 for the gating strategy).
Single-genome amplification sequencing
Viral RNA was isolated from plasma using a QIAamp Viral RNA kit (Qiagen). Extracted RNA was reverse transcribed into complementary DNA (cDNA) using SuperScript III (Qiagen) and viral-specific primer nFL-R1 (5′-CACTAGCTTACTTCTAAAATGGCAGC-3′). The cDNA was then diluted to a single-genome template before PCR with nFL-R1 and nFL-F1 (5′-GATTGGCGCCYGAACAGGGACTTG-3′) primers. Second-round PCR was performed with nFL-R2 (5′-TACTTCTAAAATGGCAGCTTTATTGAA-3′) and nFL-F2 (5′-GTGAAGGCAGTAAGGGCGGCAGG-3′) primers. Both PCR reactions were amplified with Platinum SuperFi DNA polymerase (Thermo Fisher Scientific), and correct-sized amplicons were directly sequenced using BigDye Terminator Sanger sequencing (Thermo Fisher Scientific) with multiple virus-specific primers. Sequences were aligned using Geneious Prime v.2023.0.
Env MiSeq
An Illumina-based sequencing approach was implemented on a MiSeq instrument to query the a4b7 binding site and V2V3 regions of the viral genome, as previously described48. We generated cDNA as described above with the virus-specific primer a4b7.cDNA (5′-TTCTGCCACCTCTGCACTCATGG-3′). MiSeq PCR was performed using a4b7.P7 (5′-TAGAACTTATATTTACTGGCATGG-3′) and a4b7.P5 (5′-GCCACCTCTGCACTCATGG-3′) primers. cDNA for the V2V3 region was generated using the V2.V3.cDNA primer (5′-CCTATCATTGATTGGTTGTGAGTG-3′), and PCR was conducted using V2.V3.P7 (5′-CAGGATAATTGCACAGGCTTGG-3′) and V2.V3.P5 (5′-GCCACCTCTGCACTCATGG-3′).
Statistical analysis
Data were analysed and plotted using JMP 15 and GraphPad Prism v.10.0.3 with statistical tests as detailed in figure legends or text. P < 0.05 was considered to indicate statistical significance (adjusted for multiple comparisons as appropriate).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.