Peptide synthesis
Detailed synthetic methods and characterization of all peptides, pooled peptide libraries, custom peptide building blocks and protein–peptide conjugates generated for this study are provided in the Supplementary Information. Peptide sequences are provided in Supplementary Table 9.
General method for the preparation of peptides for sortase reaction
Amino acids were loaded on chlorotrityl resin to access C-terminal carboxylic acids or Rink amide to access C-terminal amides (Supplementary Information). Automated peptide elongation was carried out on a Multisyntech Syro I parallel synthesizer according to the general peptide methods (Supplementary Information). The peptide was cleaved from the resin using TFA/DODT/H2O (95:2.5:2.5, v/v) for 1 h. The resin was removed by filtration and the filtrate concentrated under reduced pressure. The solution was triturated with diethyl ether and centrifuged to obtain crude peptide. The crude peptide was dissolved in H2O/CH3N (1:1, v/v) + 0.1% (v/v) TFA and purified using preparative high-performance liquid chromatography. The peptide series, including pep_AARS1, pep_lib_VIN and pep_lib_YNR, was obtained from Craftide.
Sortase-mediated protein conjugation
Peptides were dissolved in sortase reaction buffer (50 mM Tris, 150 mM NaCl, pH 7.4, at 4 °C). The pH was adjusted to 7–8. Peptide (final concentration 1 mM) was mixed with reporter protein (sfGFP, mCherry, BFP) (final concentration 75 µM). SortA 7 M was added (final concentration, 2 µM) and incubated at 4 °C for 18 h. Unreacted reporter protein and cleaved sortag were removed by Ni-NTA purification. The flow-through was collected and immediately buffer-exchanged to ion-exchange buffer (25 mM Tris, pH 8.5) using a desalting column (Cytiva). The sample was further purified by anion exchange using a MonoQ column (Cytiva) with a gradient of 0–25% high-salt buffer (25 mM Tris, 1 M NaCl, pH 8.5) in 25 CV. The fractions containing the product were pooled, buffer-exchanged to sortase reaction buffer supplemented with 0.5 mM TCEP and concentrated.
General cell culture methods
HEK293 and HEK293T cells were cultured in DMEM containing GlutaMax (Thermo Fisher Scientific) and K562 cells were cultured in RPMI 1640 containing GlutaMax (Thermo Fisher Scientific) under standard conditions. Growth medium was further supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific) and 100 U ml−1 penicillin–streptomycin (Thermo Fisher Scientific). Stably transduced cells were selected using 2 µg ml−1 puromycin (Thermo Fisher Scientific), 500 µg ml−1 geneticin (Thermo Fisher Scientific) or 10 µg ml−1 blasticidin (Thermo Fisher Scientific) starting 2 days after transduction.
Induction of doxycycline-dependent vectors was performed by addition of 500 ng ml−1 doxycycline (Merck) every 48 h. Cells were treated with epoxomicin (Merck), folimycin A (concanamycin A, Merck), TAK243 (MedChem Express) or MLN4924 (MedChem Express), CSN5i-3 (MedChem Express), carfilzomib (MedChem Express), menadione (Merck) and auranofin (MedChem Express) or CB-5083 (MedChem Express) as indicated in the figure legends. H2O2 was freshly diluted in serum-free DMEM growth medium to 200 µM immediately before use and was kept protected from direct light. Cell lines were regularly tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza).
All of the cell lines were obtained from certified vendors. Human cell lines were regularly tested for mycoplasma contamination using a commercially available kit (Lonza, MycoAlert). Throughout the duration of this study, no contamination was detected. CRISPR-competent K562 clones were reauthenticated by STR genotyping (Microsynth).
Culture, editing and differentiation of NPCs
Human NPCs were grown from a previously established culture derived from the H9 human embryonic stem cell line49. Cells were expanded on plates coated with laminin (R&D Systems, 3446-005-01) in NPC base medium (50% Neurobasal Plus Medium (A3582901), 50% DMEM/F-12 (31331028), supplemented with 1× GlutaMax, 35050061, 1× N-2 supplement (17502048), 1× B-27 supplement without vitamin A (12587010) and 50 µM 2-mercaptoethanol (31350010); all Thermo Fisher Scientific). The medium was freshly supplemented with 10 ng ml−1 humanKine bFGF (Merck, GF446-50UG) or 10 ng ml−1 heat stable recombinant human FGF-basic (PeproTech, 100-18HS), 10 ng ml−1 EGF (PeproTech, AF-100-15) and 20 ng ml−1 BDNF (PeproTech, 450-02).
For neuronal differentiation, cells were seeded onto plates coated with laminin and polyornithine (Merck, P3655). Differentiation was induced by gradual withdrawal of growth factors (twofold dilution in growth-factor-free NPC medium on days 1, 3 and 5) followed by a serial twofold dilution of NPC base medium in neuronal maturation medium (Neurobasal plus medium, supplemented with GlutaMax, full B-27 supplement (Thermo Fisher Scientific, A3582801), MEM non-essential amino acids (Thermo Fisher Scientific, 11140050), antibiotic-antimycotic (Thermo Fisher Scientific, 15240062), dibutyryl cyclic AMP (StemCell Technologies, 100-0244), 20 ng ml−1 BDNF and 20 ng ml−1 GDNF (Peprotech, 450-10)) on days 7 and 11. From day 13 on, cells were grown in full neuronal maturation medium until collection.
CRISPRi-competent NPCs were generated by knock-in of a modified dCas9-KRAB-BFP cassette into the CLYBL locus according to a previously established strategy for stable CRISPRi in mature human neurons50. NPCs were electroporated using the Amaxa Nucleofector II and the Cell Line Nucleofector Kit V (Lonza) to deliver TALEN plasmids for CLYBL targeting51 (pZT-C13-L1, pZT-C13-R1; Addgene, 62196 and 62197) and the homology donor pJC2528 (Supplementary Table 9). Cells were selected for correct integration using G418 (0.1 mg ml−1, Thermo Fisher Scientific). Positive clones were pooled and expanded. To activate dCas9-KRAB expression, cells were electroporated with pCAG-Cre52 (Addgene, 13775) and BFP-expressing cells were enriched by fluorescence-activated cell sorting at 72 h after electroporation.
Virus production
Lentiviral vectors were packaged in HEK293T cells using standard methods. In brief, cells were incubated with plasmid DNA (transfer plasmid, pCMV-dR8.2 dvpr and pCMV-VSV-G at a weight ratio of 4:2:1) and polyethyleneimine (PEI, molecular mass of around 25,000 Da) at a ratio of 1:3 (w/w) of total plasmid DNA to PEI. The viral supernatant was collected at 48–72 h after nucleofection by ultrafiltration and supplemented with 4 µg ml−1 polybrene. Gene knockdown was performed in a subclonal K562 cell line stably expressing dCas9-KRAB-BFP (Addgene, 102244), provided by E. J. Aird. For CRISPRi, sgRNAs targeting near transcription start sites were chosen based on previously optimized design rules53 and cloned into the vector pCRISPRia. Puromycin was used to select for cells stably expressing sgRNAs. All custom vectors generated for this study will be made available through Addgene after publication of this Article. A complete list of plasmids used in this study is provided in Supplementary Table 9.
Flow cytometry assays and cell sorting
Analytical flow cytometry was performed on the Attune NxT flow cytometer (Thermo Fisher Scientific). Single-cell isolation and sorting for CRISPR-screen analysis was performed on the Sony SH-800 cell sorter. Flow cytometry data were quantified and visualized using FlowJo 10.
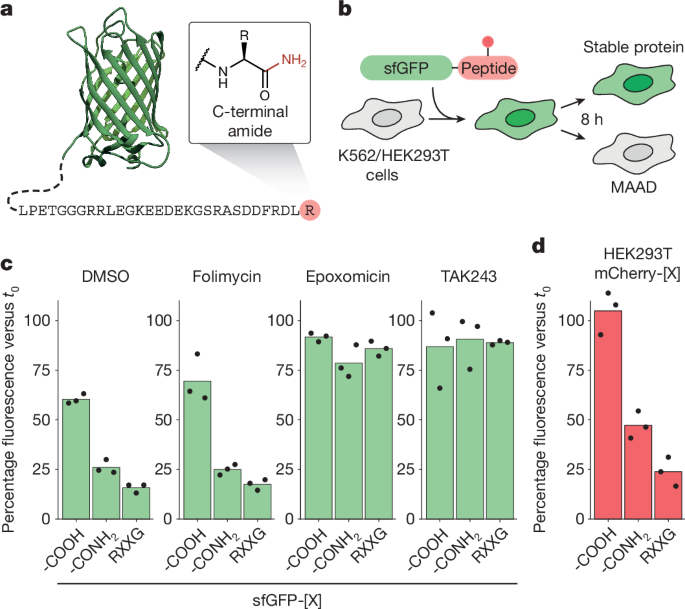
For measuring the stability of recombinant or semi-synthetic proteins, 1.5–2.0 × 105 cells were electroporated with 200 pmol of protein in 20 µl cuvettes using a 4D nucleofector device (Lonza) according to the manufacturer’s recommendations. After protein delivery, cells were left to recover at 37 °C for 30–45 min and washed with PBS before measuring the initial mean fluorescence intensity values (t0) using flow cytometry. Subsequent measurements were performed at the indicated timepoints. Baseline fluorescence values of mock-electroporated cells receiving no protein were measured in parallel and subtracted for each timepoint. Background-corrected fluorescence values were normalized to t0.
Cell surface expression of CD55 was assessed by immunostaining and flow cytometry. In brief, cells were pelleted and resuspended in primary antibody diluted 1:200 in PBS containing 5% FBS. (APC-anti-human-CD55[JS11], BioLegend). After 20 min of incubation on ice, cells were washed three times with PBS followed by data acquisition. For competitive growth assays, cells were transduced with lentiviral vectors delivering FBXO31-IRES-GFP cDNA variants or an empty vector control (IRES-GFP) at a multiplicity of infection (MOI) of about 0.3. The fraction of transduced cells was measured every 2 days by flow cytometry and normalized to initial transduction levels (day 2) as a measure of relative fitness.
For mapping FBXO31(D334N) neosubstrate degrons, we cloned sequences encoding the C-terminal 15 amino acids of candidate substrates or mutants thereof into the C terminus of GFP in the previously established two-colour reporter vector Cilantro240 (Supplementary Table 9). After G418 selection and sorting of transduced cells, FBXO31 cDNAs were delivered by transduction with lentiviral vectors encoding for FBXO31-IRES-mTagBFP or its D334N-mutant version (Supplementary Table 9). Fluorescence of the GFP reporter and mCherry control was measured by flow cytometry 2 days after transduction. Protein stability among mTagBFP-positive cells was measured as the ratio of median fluorescence values of GFP and mCherry (Supplementary Fig. 1d).
Generation of clonal cell lines
For genome-wide CRISPR screening, K562 cells were made competent for inducible gene knockout (iCas9) by transduction with vectors SRPB (pHR-SFFV-rtTA3-PGK-Bsr) and 3GCasT (pHR-TRE3G-hSpCas9-NLS-FLAG-2A-Thy1.1). Cas9-P2A-Thy1.1 expression was induced with doxycycline for 2 days and cells staining positive for Thy1.1 were isolated by single-cell sorting. Clonal lines were screened for cells that show no evidence of CD55 knockout after viral delivery of sgCD55 (Supplementary Table 9) by antibody staining and flow cytometry, as well as efficient knockout after addition of doxycycline for 9 days.
FBXO31-knockout cells were generated by electroporation of cells with Cas9–sgRNA ribonucleoprotein particles as described previously54 using the sgRNAs listed in Supplementary Table 9 based on previously optimized design rules20,55. In brief, in vitro transcription templates were generated by PCR using Q5 polymerase (New England Biolabs) with the primers listed in Supplementary Table 9 and used for in vitro transcription by T7 RNA polymerase (New England Biolabs). The resulting RNA was purified using a spin-column kit (RNeasy mini kit, QIAGEN) and 120 pmol of sgRNA was complexed with 100 pmol of recombinant SpCas9 protein at room temperature for 20 min. Cas9 protein was provided by the Genome Engineering and Measurement Lab (GEML) of the Functional Genomics Center Zurich (FGCZ of the University of Zurich and ETH). Assembled sgRNA–Cas9 complexes were delivered to cells using the 4D nucleofector kit (Lonza) according to the manufacturer’s instructions. Clonal cell lines were isolated by single-cell sorting and characterized by genomic DNA extraction (Lucigen QuickExtract), PCR amplification of edited loci using Q5 polymerase (New England Biolabs) with genotyping and NGS primers (Supplementary Table 9). Pooled next-generation sequencing of edited loci was performed by GEML (FGCZ of the University of Zurich and ETH) on the MiSeq sequencer (Illumina) with 150 bp paired-end reads. Deep sequencing reads were analysed using CRISPResso256 (v.2.3.0) to identify compound heterozygous knockout clones.
Western blotting and antibodies used in this study
Except for co-IP studies, immunoblotting was performed on whole-cell lysates in radioimmunoprecipitation buffer (RIPA, 50 mM Tris-HCl, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, pH 7.4) supplemented with protease inhibitors (Halt protease inhibitor cocktail, Thermo Fisher Scientific). Proteins were analysed by polyacrylamide gel electrophoresis (PAGE) and wet transfer onto nitrocellulose membranes (0.2 µm pore size) using standard methods. Membranes were blocked with TBS-T (150 mM NaCl, 20 mM Tris, 0.1% (w/v) Tween-20, pH 7.4) containing 5% skimmed milk powder and incubated with primary antibodies diluted in TBST containing 5% bovine serum albumin (BSA) and 0.05% (w/v) sodium azide. The primary western blotting antibodies used in this study were as follows: FBXO31 (Abcam, ab86137; Human Protein Atlas, HPA030150), GFP (Abcam, ab6556), mCherry (Abcam, ab183628), SKP1 (Cell Signaling Technology, 2156), CUL1 (Invitrogen, 71-8700), AARS1 (Fortis/Bethyl Life Science, A303-473A), GLUL (Fortis/Bethyl Life Science, A305-323A), cyclin D1 (Abcam, ab134175), HA (Cell Signaling Technology, 3724) and SUGT1 (Bethyl, A302-944A). Primary western blotting antibodies for mCherry and GFP were used at a dilution of 1:2,000. All of the other primary western blotting antibodies were used at a 1:1,000 dilution. Detection of primary antibodies was performed using far-red fluorescently labelled secondary antibodies (LI-COR Biosciences, 926-32213 and 926-68072) at a dilution of 1:15,000. The blots were scanned on the Odyssey CLx scanner (LI-COR Biosciences).
CRISPR screening and analysis
iCas9 cells were transduced with the pooled lentiviral sgRNA library TKOv320 at an MOI of about 0.3 as measured by serial dilution, puromycin selection and viability assay (CellTiter-Glo 2.0, Promega). Two pools of 1.2 × 108 cells each were transduced, yielding an approximately 500-fold library coverage, which was maintained throughout all of the cell culture steps. Cas9 expression was induced by addition of doxycycline for 5 days before delivery of reporter proteins. To isolate CTAP-clearance-deficient cells., ≥4 large-scale nucleofections were performed by combining 5 × 107 cells, 2,000 pmol sfGFP-pep1-CONH2 (sfGFP-GGGKDLEGKGGSAGSGSAGGSKYPYDVPDYAKS-[CONH2]) and 2,000 pmol of a degron-tagged control protein (mTagBFP2–RXXG) in a 100 µl nucleofection reaction (4D nucleofector kit SE plus supplement SF1, Lonza)
Next, 14 h after nucleofection, cells were transferred on ice and sorted into a CTAP-clearance-deficient population (sfGFP+ mTagBFP2−) and a control population (sfGFP−mTagBFP−). Genomic DNA was extracted from snap-frozen sorted cells using the Gentra Pure kit (QIAGEN). sgRNA cassettes were isolated by two rounds of PCR using a previously published strategy20 with NEBNext Ultra II Q5 Master Mix (New England Biolabs) and primer pairs listed in Supplementary Table 9.
Protospacers were quantified by deep sequencing using the NovaSeq SP 200 platform (Illumina) with 21 initial dark cycles by GEML (FGCZ of the University of Zurich and ETH) . sgRNA counts were retrieved using ‘mageck count’ (MAGeCK v.0.5.9.3) with the default parameters57. Raw sgRNA counts are provided in Supplementary Table 1. Enrichment of sgRNAs targeting the same gene in CTAP-clearance-deficient cells versus the control population was estimated with the ‘mageck test’ command using a paired design for screening duplicates with option –remove-zero both and otherwise the default parameters.
Recombinant protein production in Escherichia coli
Fluorescent protein expression and purification: sortase-tagged variants of GFP, mCherry and mTagBFP2 were expressed from IPTG-inducible expression plasmids listed in Supplementary Table 9 in E. coli strain BL21(DE3). Expression was induced at an optical density at 600 nm of 0.6 using 1 mM IPTG followed by continued culture for 6 h at 37 °C and freezing of cell pellets. The samples were lysed by sonication in buffer NBA (200 mM NaCl, 50 mM HEPES, 25 mM imidazole, 1 mM TCEP, pH 8.0) His-tagged protein was enriched from cleared lysates on the HisTrap FF column (Cytiva) by elution with 250 mM imidazole in sonication buffer. Crude HisTrap eluates were used as input for sortylation followed by further purification as described above.
Codon-optimized cDNAs of SKP1(A2P,∆38–43,∆71–82) and His6-Smt3-StrepII-FBXO31.1(∆1–65) were generated by gene synthesis (Integrated DNA Technologies) based on previously optimized expression constructs58,59 and cloned into the bacterial expression vector pET28b (Supplementary Table 9). Expression was performed at 18 °C overnight in the presence of 0.5 mM IPTG in E. coli strain BL21(DE3) in Terrific Broth (Faust) in the presence of 50 µg ml−1 kanamycin. After bacterial cell lysis by sonication in buffer NBA (200 mM NaCl, 50 mM HEPES, 25 mM imidazole, 1 mM TCEP, pH 8.0), recombinant protein was enriched on the HisTrap FF column (Cytiva) by elution with 250 mM imidazole in the same buffer. The His6–Smt3 tag was removed by digestion with purified SUMO protease Ulp1 at 4 °C overnight. Binary SKP1–FBXO31 complexes were further purified by anion-exchange chromatography (HisTrap Q HP, Cytiva) in 50 mM HEPES (pH 7.5) and 5 mM DTT on a linear gradient of 100–1,000 mM NaCl followed by gel filtration (HiPrep Sephacryl S-100 HR, Cytiva) in 200 mM NaCl, 50 mM HEPES and 1 mM TCEP. Sortase (SortA-7M) and Ulp1 were prepared by recombinant expression E. coli as previously reported60,61.
Protein expression and purification in insect cells
Human cDNA sequences of tRNA-LC subunits were subcloned into modified pLIB vectors. HSPC117 was N-terminally 8×His-tagged and FAM98A(1–340) was modified with N-terminal 2×StrepII-tag. DDX1 and RTRAF (CGI-99) were subcloned without a tag and in full length. The resulting gene cassettes were combined into a single multigene construct using the bigBAC system62. The multigene construct was integrated into a baculovirus genome, transfected into SF9 cells for virus production and finally used for infection of large-scale protein production cultures as described previously63. SF9 cells were grown in SF-4 Baculo Express ICM ready to use medium (Bioconcept, 9-00F38-K). Initial transfection was performed using the TransIT-Insect Transfection Reagent (Mirus, Cat. MIR 6104). Then, 50 ml of infected preculture was used to infect 500–750 ml SF9 cultures at 1.5 million cells per ml or multiples thereof. Infected large-scale cultures were grown for 72 h at 27 °C and 90 rpm.
Large-scale cultures were pelleted by centrifugation, washed in tRNA-LC lysis buffer (500 mM KCl, 20 mM HEPES-KOH, 30 mM imidazole, pH 7.8). Flash-frozen cell pellets were stored at −80 °C. Cells were lysed in tRNA-LC lysis buffer by sonication (Bandelin Sonopuls HD 3200, VS 70T probe). Cleared lysate was incubated with equilibrated HIS-Select nickel affinity resin (Merck, P6611) at 4 °C. The resin was washed extensively with tRNA-LC lysis buffer on a gravity-flow column and captured protein was eluted in Ni-elution buffer (150 mM KCl, 20 mM HEPES-KOH, 250 mM Imidazole, pH 7.8). A second affinity purification was performed with equilibrated Streptactin Sepharose high performance resin (Cytiva, 28-9355-99) by binding for 1 h at 4 °C on a rolling table, washing on a gravity flow column with tRNA-LC wash buffer (150 mM KCl, 20 mM HEPES-KOH, pH 7.8) and eluted in Strep-elution buffer (150 mM KCl, 20 mM HEPES-KOH, 5 mM desthiobiotin, pH 7.8). The quality and efficiency of all protein purification steps was monitored using SDS–PAGE.
Pooled library IP and TIMS-TOF analysis
For each IP, 1 pmol of peptide library was incubated with an equimolar amount of recombinant Strep-II-tagged FBXO31–SKP1 complex in 1 ml of binding buffer (200 mM NaCl, 25 mM HEPES pH 7.5, 1 mM TCEP) with pre-equilibrated magnetic Strep-Tactin XT beads (240 µl MagStrep type 3 XT bead slurry, IBA life sciences). After 1 h of binding at room temperature on a rotator, the beads were washed twice with binding buffer, twice with wash buffer (150 mM NaCl, 25 mM HEPES, 1 mM TCEP) and eluted in 150 mM NaCl, 50 mM HEPES, 100 mM biotin (pH 7.7). Eluted peptides and an equal amount of input library (1 pmol) were diluted 1:1 with triethylammonium bicarbonate (50 mM, pH 8.0). Peptides were isobarically labelled with TMT 2-plex (Thermo Fisher Scientific) labels according to the manufacturer’s instructions. The input library was modified with TMT-126 and eluted peptides with TMT-127. After labelling, peptides were combined 1:1. Combined peptides were desalted using 100 µl C18 ZipTips and dried (Savant SpeedVac). The dried peptides were resuspended in 5 vol% acetonitrile, 0.1 vol% formic acid.
Tandem MS experiments were performed using the ESI-TIMS-QTOF-MS system (TimsTOF Pro, Bruker Daltonics) with collision-induced dissociation and N2 as the collision gas. Peptides were pressure loaded onto a reversed-phase 25 cm × 75 µm inner diameter C18 1.6 μm column (Ionoptics) with a reversed-phase 5 mm × 0.3 mm inner diameter C18 5 μm column (Thermo Fisher Scientific) as a guard column at 40 °C. The mobile phase consisted of water with 0.1% formic acid (A) and acetonitrile with 0.1% formic acid (B). The gradient started at 2% of B and was linearly increased to 35% B in 120 min at flow rate of 300 µl min−1. A second gradient profile, started at 35% of B and was linearly increased to 95% B in 2 min at flow rate of 300 µl min−1. This was followed by isocratic conditions of 95% B at flow rate of 300 µl min−1 for 8 min. The total run time, including the conditioning of the column to the initial conditions, was 163 min. Further data processing was performed using Data Analysis 5.3 software (Bruker Daltonics) using a processing script to generate export files and reports.
Mascot Server v.2.8.1. (Matrix Science) was used to match spectra against a custom reference of common contaminants concatenated with all potential peptides generated by pooled SPPS (10 ppm peptide mass tolerance, 0.05 Da fragment mass tolerance and, for peptide–amide libraries, −0.98 Da at peptide termini as a variable modification). For each peptide, reporter ion intensities were summed across multiple detections and scaled to the sum of reporter ion intensities for all input library peptides yielding relative reporter ion intensities for cross-comparisons between runs. For each experiment, two parallel IPs were analysed. For wild-type FBXO31 co-IP, two MS measurements were run for each IP for improved library coverage.
In vitro ubiquitylation assay
2 µM of recombinant FBXO31–SKP1 complexes was combined with 0.5 µM of substrate, 50 µM recombinant ubiquitin, 50 nM UBE1, 500 nM UBE2R1, 500 nM UBE2D3 and 500 nM CUL1–NEDD8–RBX1 complex in 50 mM Tris/HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 10 mM ATP and 0.5 mM TCEP. The reactions were incubated at 30 °C for 1 h unless indicated otherwise and analysed by immunoblotting. All protein components except for FBXO31–SKP1 and model substrates were sourced commercially (R&D Systems).
HA–FBXO31 co-IP
Co-IP of semi-synthetic model substrates (mCherry-GGGRRLEGKEEDEKGSRASDDFRDLR-[COOH/CONH2]) was performed in FBXO31-knockout HEK293 cells stably transduced with pLenti-EF1A-HA-FBXO31-PGK-Neo or the indicated mutant derivatives (Supplementary Table 9). Cells were nucleofected 2 h before collection and treated with 2 µM MLN4924 (MedChemExpress) and 500 nM epoxomicin (Merck) or 1 µM carfilzomib (MedChemExpress) to block protein degradation and stabilize CRL complexes.
Co-IP of endogenous FBXO31 clients and model CTAPs was performed in FBXO31-knockout HEK293 or HEK293T cells stably transduced with the lentiviral vectors expressing cDNAs of HA-tagged FBXO31 or mutant derivatives thereof as noted in figure legends (Supplementary Table 9). Cells were cultured in serum-free DMEM supplemented with 500 nM epoxomicin with or without addition of H2O2 at a final concentration of 200 µM for 1 h before cell collection.
For both endogenous and synthetic client IP, cells were collected by centrifugation and washed in ice-cold PBS followed by lysis in SCF-IP buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 0.1% Igepal CA-630, 5% (v/v) glycerol, pH 7.5) supplemented with Halt protease inhibitor cocktail (Thermo Fisher Scientific) by rotation for 30 min at 4 °C. Debris was removed by centrifugation (4 °C, 30 min, 20,000g) and the total protein concentrations were measured using the Bradford protein assay (Thermo Fisher Scientific). Typically, 140 µg of protein was incubated with 12 µl of Pierce anti-HA magnetic bead resin (Thermo Fisher Scientific) on a rotator wheel at 4 °C overnight. The beads were washed three times for 5 min in SCF-IP buffer and bound proteins were eluted twice using 0.1 M glycine (pH 2), followed by addition of 0.25 volumes neutralization buffer (1.5 M NaCl, 0.5 M Tris-HCl, pH 8.0).
Co-IP MS of HA-tagged FBXO31
IP–MS of HA-tagged FBXO31 was performed with FBXO31-knockout HEK293T cells stably expressing HA–FBXO31(ΔF-box) from vector pLenti-EF1A-HA-FBXO31(ΔF-box)-IRES-GFP or an indicated mutant cDNA. For each condition, three replicate samples were processed from separately passaged cultures. As a negative control, we used identical FBXO31-knockout HEK293T cells not expressing bait cDNAs. For IP analysis of oxidatively stressed cells, cultures were incubated in serum-free DMEM supplemented with 500 nM epoxomicin with or without addition of H2O2 at a final concentration of 200 µM for 1 h before to cell collection.
Cells were trypsinized and washed twice with ice-cold PBS containing 500 nM epoxomicin and resuspended in lysis buffer (300 mM NaCl, 50 mM Tris-HCl, 0.5% (v/v) Igepal CA-630) freshly supplemented with Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). Cell lysates were obtained by rotation for 30 min at 4 °C and were cleared by centrifugation (4 °C, 30 min, 20,000g). For IP, 1,000 µg of cellular protein extract was incubated with 80 µl of pre-equilibrated bead slurry (Pierce anti-HA magnetic beads, Thermo Fisher Scientific) in a total volume of 250 µl of lysis buffer on a rotator wheel at 4 °C for 4 h. Magnetic beads were washed four times with lysis buffer and twice with PBS followed by elution with 80 µl 0.1 M glycine (pH 2.0) at room temperature with gentle shaking for 10 min. The supernatant was neutralized with 0.25 volumes of 1.5 M NaCl, 0.5 M Tris-HCl pH 7.5.
Eluates were processed by the proteomics group of FGCZ (University of Zurich and ETH) by in-solution tryptic digest and LC–MS. In brief, LC–MS analysis was performed on the Orbitrap Exploris 480 Mass Spectrometer (Thermo Fisher Scientific) coupled by ESI to the ACQUITY UPLC M-Class System (Waters). Data-dependent acquisition was performed over a 65 min runtime with MS1 precursor mass scans at 120,000 resolution across 350–1,200 m/z. Precursors were isolated with a 1.2 m/z window followed by HCD fragmentation at 30% collision energy. MS2 scans were performed at 30,000 resolution across 350–1,200 m/z.
Spectra were searched against the human proteome (UniProt: UP000005640) using the MSFragger engine64,65 (v.4.1) through FragPipe (v.22.0) with the default parameters for closed search of fully tryptic peptides and sequential FDR filtering of peptide spectrum matches (PSMs), peptides and proteins by philosopher66 with the default parameters (protein FDR ≤ 0.01). Common contaminants67 were removed and enrichment of interactors was calculated using limma v.3.58.1, by fitting a linear model to normalized spectral counts for each sample plus one pseudocount followed by empirical bayes moderation and Benjamini–Hochberg correction of P values for multiple-hypothesis testing. Detailed search parameters and acquisition settings are deposited alongside the raw data at Pride (https://www.ebi.ac.uk/pride/).
IP–MS of Flag-tagged AARS1
IP–MS analysis of 3×Flag-tagged AARS1 was performed on FBXO31-knockout HEK293T cells transiently transfected with pCDNA4TO-CMV-3xFLAG-Halo-AARS1 using 30 µg of PEI (molecular mass, ~25,000) and 10 µg of plasmid DNA. Two days after transfection, cells were challenged with 200 µM H2O2 in serum-free DMEM. Then, 5 min after H2O2 treatment, cells were additionally supplemented with 500 nM epoxomicin. Cells were washed three times in PBS supplemented with 500 nM epoxomicin and snap-frozen. Cell pellets were lysed by resuspension in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, pH 7.4) supplemented with protease inhibitors (Halt protease inhibitor cocktail, Thermo Fisher Scientific). Then, 250 µg of protein was incubated with 25 µl of pre-equilibrated anti-Flag resin (M2 anti-FLAG beads, Merck) on a rotator at 4 °C overnight. The beads were washed once with RIPA buffer, four times with high-salt RIPA buffer (50 mM Tris-HCl, 500 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, pH 7.4) and twice with TBS (150 mM NaCl, 50 mM Tris pH 7.5) followed by elution in 100 µl TBS + 150 ng µl−1 competitor peptide (Pierce 3× DYKDDDDK Peptide, Thermo Fisher Scientific) at 37 °C for 30 min. The eluates were processed by the proteomics group of FGCZ (Uni Zürich/ETH) by in-solution digest with Glu-C protease and LC–MS/MS in data-dependent acquisition mode on the Orbitrap Exploris 480 Mass Spectrometer as described above for IP–MS analysis of FBXO31.
Spectra were searched against the human proteome (UniProt: UP000005640) using the MSFragger engine64,65 (v.4.1) through FragPipe (v.22.0) with the default parameters for semi-tryptic search unless stated otherwise. In brief, peptide identification was based on b– and y-series ions allowing for up to 1 missed cleavage and up to 1 non-enzymatic peptide terminus, variable peptide C-terminal amidation (−0.984016 Da), M oxidation (+15.99490 Da), protein C-terminal acetylation (+42.01060 Da) and fixed C carbamidomethylation (+57.021464 Da). Sequential filtering for high-confidence PSMs, peptides and proteins was performed using philosopher66 with the default parameters (protein FDR ≤ 0.01). For identification of amide-bearing de novo C termini, internal AARS1-matching peptides were filtered for modified C termini not matching a Glu-C cleavage site (D or E). Detailed search parameters are deposited alongside the raw data at Pride (https://www.ebi.ac.uk/pride/).
Expression proteomics by tandem mass tag MS
For inducible expression of DD-3×Flag-FBXO31, FBXO31-knockout HEK293 cells were transduced with pLenti-EF1A-DD-3×Flag-FBXO31-PGK-Neo or its D334N mutated variant (Supplementary Table 9). Cells were treated with 2 µM Shield-1 (MedChem Express) for 12 h before collection. All of the experiments were performed with three replicate samples treated and collected on separate days. Cell pellets were processed for whole-proteome quantification by FGCZ (Univesity of Zurich and ETH). In brief, cell pellets were lysed in 4% SDS in 100 mM Tris/HCl pH 8.2 by boiling and mechanical homogenization. Per sample, 50 µg of total protein was reduced (2 mM TCEP) and alkylated (15 mM 2-chlroacetamide) and further processed using a fully automated SP3 purification, digest and clean-up workflow68. The samples were isobarically labelled with TMT10plex (Thermo Fisher Scientific), pre-fractionated by RP-HPLC on an XBridge Peptide BEH C18 column (Waters) and pooled into eight fractions.
In brief, LC–MS analysis was performed on the Orbitrap Exploris 480 Mass Spectrometer (Thermo Fisher Scientific) coupled by ESI to the ACQUITY UPLC M-Class System (Waters). Data-dependent acquisition was performed over 110 min runtime with MS1 precursor mass scans at 120,000 resolution across 350–1,400 m/z. Precursors were isolated with a 1.2 m/z window followed by HCD fragmentation at 30% collision energy. MS2 scans were performed at 45,000 resolution across 350–1,400 m/z.
Raw MS data were processed using ProteomeDiscoverer 2.4 (Thermo Fisher Scientific). Spectra were searched against the UniProt human reference proteome (downloaded on 29 April 2022) and common contaminants with Sequest HT with a peptide-level FDR cut-off of 0.01. Detection of differentially abundant proteins was performed on reporter ion intensities for D334N or wild-type FBXO31-expressing cells compared with Shield-1-treated control cells lacking FBXO31 cDNA.
Re-analysis of public proteomes for CTAP detection
Label-free DDA-MS-based proteomics of tryptic digests of healthy human tissues from single donors were downloaded from PRIDE archive PXD010154, samples 1277 (brain), 1499 (adipose tissue), 1306 (thyroid) and 1296 (heart)69. Peptide search was performed exactly as described above for Flag–AARS1 IP–MS. For downstream analysis, fully enzymatic peptides were defined as having termini that match trypsin cleavage sites or annotated N and C termini, accounting for clipping of N-terminal Met. Semi-enzymatic peptides were defined as having one cleavage that does not match an annotated protein terminus or trypsin cleavage site. Full search parameters and PSM-level search results used for quantification of CTAP formation are deposited at Pride (https://www.ebi.ac.uk/pride/).
In vitro protein fragmentation monitoring
Purified human haemoglobin was obtained from Abcam (ab77858) and dissolved in DPBS (Thermo Fisher Scientific 14190144). In vitro fragmentation reactions were performed at a final concentration of 1 g l−1 haemoglobin and indicated concentrations of H2O2 by incubation at 37 °C for 1 h. Purified human tRNA-LC was diluted in PBS to a final reaction volume of 0.25 g l−1 and incubated with the indicated concentrations of H2O2 and CuCl2 for 30 min at room temperature for in vitro fragmentation. All of the reactions were stopped by flash-freezing in liquid nitrogen. Thawed reactions were immediately precipitated and processed for tandem MS by the FGCZ (University of Zurich and ETH). In brief, proteins were precipitated by addition of TCA (Merck) to a final concentration of 5%, washed twice with ice-cold acetone, air-dried and resuspended in aqueous buffer containing 10 mM Tris (pH 8.2) and 20 mM CaCl2. The samples were reduced and alkylated with 5 mM TCEP and 15 mM 2-chloroacetamide (30 min, 30 °C) followed by enzymatic digest with the indicated enzymes (trypsin, ArgC, trypsin-N or chymotrypsin). LC coupled to MS in data-dependent acquisition mode was performed as described above for FBXO31 IP–MS experiments.
Raw MS/MS files were searched for semi-enzymatic peptides using the MSFragger engine64,65 (v.4.1) through FragPipe (v.22.0) with the default parameters for the respective proteases additionally allowing for peptide-C-terminal amidation (−0.9840 Da). Search was performed against the respective purified proteins and common contaminants with decoys. Sequential filtering was performed with Philosopher for protein FDR < 0.01 (v.5.1.1). In addition to initial filtering, only PSMs with a probability of >0.01 were used for plotting. Complete search parameters are deposited alongside raw data files and search results at the PRIDE archive (https://www.ebi.ac.uk/pride/).
As a control for technical noise, in vitro fragmented haemoglobin was reanalysed also allowing multiples of 0.984 as C-terminal mass shifts (from −2.952 to +2.952).
Active CRL profiling
Treatments were performed in four separate cultures as individual replicates for each tested condition. For active CRL profiling, a subclonal line of K562 cells was pretreated with 1 µM carfilzomib (MedChem Express, HY-10455) and 5 µM p97/VCP inhibitor CB-5083 (MedChem Express, HY-12861) for 5 min. Cells were either left untreated or treated with 200 µM freshly diluted H2O2 for 2 h. To preserve the active CRL repertoire during cell collection, cells were treated with an N8-block protocol70. Specifically, cells were exposed to 1 µM MLN4924 (MedChemExpress, HY-70062) and 1 µM CSN5i-3 (MedChemExpress, HY-112134) in PBS for 3 min. After treatment, cells were washed with PBS (Gibco, 14190094) and snap-frozen in pellets of around 5 × 106 cells each. Thawed pellets were lysed in 380 µl of lysis buffer (25 mM HEPES pH 7.4, 5% glycerol, 150 mM NaCl, 0.5% NP-40, 1× HALT protease/phosphatase inhibitor (Thermo Fisher Scientific, 78440), 2 µM MLN4924, 2 µM CSN5i-3). Lysates were clarified by centrifugation at about 20,000g for 3 min at 4 °C and filtered through 0.22 µm spin filters (Corning, 8161). An active Cullin-binding Fab31 was immobilized on high-capacity magnetic Streptavidin beads (Promega, V7820) according to the manufacturer’s instructions. The equivalent of 10 µl bead slurry of the Fab-coated beads was then added to the cleared lysates and incubated for 45 min at 4 °C with gentle rotation. Beads were washed twice with lysis buffer, twice with wash buffer (lysis buffer without IGEPAL CA-63) and twice with HBS (25 mM HEPES pH 7.4 and 150 mM NaCl). Elution was performed with 50 µl 0.1% TFA in water. The elution was then heated at 98 °C for 5 min. Subsequently, 2-chloroacetamide and TCEP were added to final concentrations of 40 mM and 10 mM, respectively, and the samples were incubated at 45 °C for 5 min. Digestion was carried out overnight at 37 °C with agitation (1,200 rpm) using a 1:100 w/w ratio of trypsin (Sigma-Aldrich, T6567) and LysC (FUJIFILM Wako, 125-05061). The samples were quenched by addition of TFA to a final concentration of 1% and 200 ng of peptides were loaded Evotips Pure (EvoSep, EV2011) according to the manufacturer’s instructions.
The samples were measured using the Evosep One LC system (EvoSep, EV-1000) coupled to a TimsTOF Pro 2 mass spectrometer (Bruker Daltonics). The 30 samples per day method was used with a 15 cm × 150 μm column packed with 1.9 μm C18-beads (Bruker Daltonics, 1893471), maintained at 50 °C and coupled to a 10 µM fused silica ID emitter (Bruker Daltonics, 1865691) within a CaptiveSpray ion source (Bruker Daltonics). Mobile phases were composed of 0.1% formic acid in water (buffer A) and 99.9% acetonitrile/0.1% formic acid (buffer B). For data acquisition, a dia-PASEF method with 20 dia-PASEF scans separated into two ion mobility windows per scan, covering an m/z range from 350 to 1,200 was used. Variable window widths were determined using py_diaid71, and the ion mobility range was set between 0.7 to 1.3 V s cm−2. The accumulation and ramp times were both set to 100 ms. Collision energy was ramped linearly from 20 eV at 1/K0 = 0.6 V s cm−2 to 59 eV at 1/K0 = 1.6 V s cm−2.
DIA raw files were searched using library-free search in DIA-NN72 (v.1.8.1, permitting one missed cleavage, one variable modification (N-terminal acetylation and methionine oxidation), cysteine carbamidomethylation as a fixed modification, with MBR and ‘deep-learning-based spectra, RT and IM prediction’ enabled). Data were analysed using the Perseus software package73 (v.1.6.7.0). Protein intensities were log2-transformed, and the datasets were filtered to contain no missing value in at least one experimental condition for all of the protein groups. Missing values were imputed using a normal distribution with a width of 0.3 and a downshift of 1.8.
Visualization of protein structures and alignments
All protein structures were downloaded from the PDB under the accession numbers noted in respective figure legends. Structures and electrostatic potential maps were rendered using ChimeraX-1.574. The composite structure of SCF–FBXO31 was generated from substructures 5VZU (FBXO31) and 6TTU (NEDD8–CUL1–RBX1–SKP1). Structures were aligned by their shared SKP1 substructure in using MatchMaker with the default settings. Alignment and conservation analysis of select FBXO31 orthologues and of FBXO31(D334N) neosubstrates was performed using JalView75 (v.2.11).
FP assays and analysis
Increasing concentrations of FBXO31–SKP1 complex (0–5 µM) were incubated with fluorescein-labelled peptides (2–10 nM) in 25 mM HEPES, 150 mM NaCl, pH 7.5 and incubated at room temperature for 30 min. FP was measured on a Victor Nivo Plate Reader (Perkin Elmer). After subtraction of the baseline signal, the polarization levels were normalized to the highest signal of each pair of amidated and non-amidated peptides for wild-type FBXO31. Binding curves were fitted by least-squares regression and half-maximal binding concentrations were extracted using Prism 9 (Graphpad) assuming one-site binding, maximal binding at 100% of the measured signal and no contribution from unspecific interactions. Dissociation constants for the highest affinity probes reported in this study are close to the probe concentrations used for FP assays and are therefore reported only as approximate values.
RNA-seq and differential gene expression analysis
Total RNA was collected from CRISPRi competent HEK293T cells, NPCs and NPC-derived neurons by column purification (RNeasy Mini, QIAGEN). Poly-A-enriched full-length mRNA-sequencing libraries were generated and sequenced by Novogene (RSPR00102) using the NovaSeq X Plus series platform for 150 bp paired-end sequencing. Raw sequencing reads were aligned to a custom human genome reference based on GrCH38 without alternative chromosomes and including commonly used cDNAs. Reads were aligned with STAR-aligner76 (v.2.7.10a) and counting was performed using FeatureCounts77 (v.2.0.6) against a custom transcriptome reference. Differential gene expression analysis was performed using DESeq278 (v.1.42.1) with the default parameters. Differentially expressed genes were searched against a database of published experimental signatures with rummagene79. Transcriptional changes in in vitro differentiated somatic motor neurons carrying amyotrophic-lateral-sclerosis-associated mutations were plotted based on previously published differential gene expression results34.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.