Animals
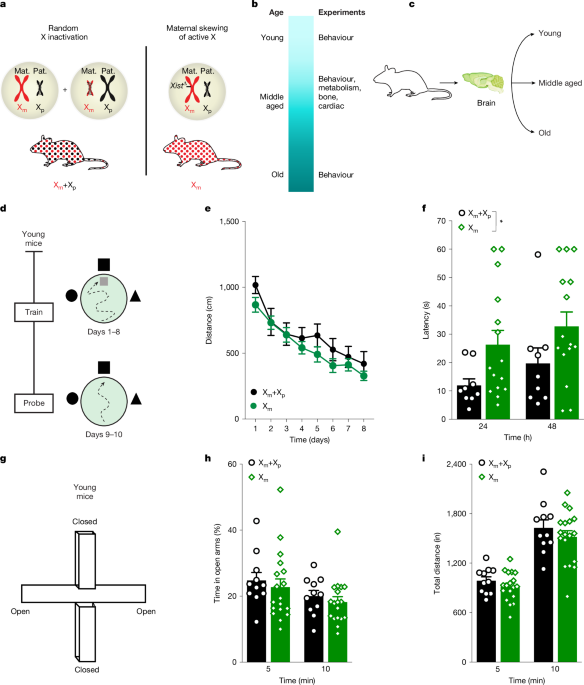
The Institutional Animal Care and Use Committee of the University of California San Francisco (UCSF) approved all animal studies. Mice were kept on a 12 h light–dark cycle, with temperatures between 19 °C and 23 °C and humidity between 30% and 70% with ad libitum access to food and water. The standard housing conditions were five mice per cage except during Morris water maze experiments and metabolic tests for the Comprehensive Lab Animal Monitoring System (CLAMS), when mice were singly housed. All experiments were carried out during the light cycle with the exception of the CLAMS metabolic tests, for which data were collected throughout the light and dark cycles. To assess effects of Xm skew compared with mosaicism, we generated mice with global maternal Xm-only expression (Xm mice) with non-transgenic littermates showing normal, random X inactivation (Xm+Xp mice). Xm-only expression was achieved by Xist deletion. Of note, Xist is a long noncoding RNA that regulates random X-chromosome inactivation. In brief, we crossed 129-Xisttm2Jae/Mmnc mice10 obtained from the Mutant Mouse Resource and Research Centers with Zp3cre mice, provided by S. Kalantry11. F2 mice were then backcrossed to C57BL/6J mice to obtain a congenic C57BL/6J background, which was verified by genetic testing. Female mice underwent multiple tests of metabolism, cardiac function, body composition, behaviour and cognition during life stages indicated in the captions of Figs. 1–4. All arenas and equipment were cleaned with 70% ethanol between tests, except for the water maze. All experimenters were blind to mouse genotypes and groups.
To assess the differences between Xm and Xp neurons from the same brain, we used well-characterized mice obtained from the laboratory of J. Nathan24 that were generated to carry X-linked, Cre-activated and nuclear fluorescent reporters of GFP on one X chromosome and tdTomato on the other. They were also backcrossed to obtain a congenic C57BL/6J background. These reporter mice possess a floxed tdTomato fluorescent protein or a floxed GFP protein inserted into the Hprt locus of the X chromosome using modified Hprt-targeting vectors. The Hprt locus is subject to random X-chromosome inactivation and inserting the fluorescent proteins in this position ensures that once crossed with a suitable Cre line, either GFP or tdTomato is expressed from each cell but never both. The Cre line used to drive cell-type-specific Xm and Xp fluorescence was well characterized with a synapsin I, neuron-specific promoter40.
Cardiac function
Measurements of cardiac function were performed as described48. In brief, mice were anaesthetized with isoflurane. Body temperature was monitored throughout the procedure using a rectal probe. A warm ultrasound gel was applied to the chest. Using a MX550S transducer, the B and M mode parasternal short-axis view was recorded, the diameter of the left ventricular lumen was measured and the ejection fraction was calculated. Afterwards, electrodes were removed, the ultrasound gel was removed and animals were allowed to recover before being returned to their cages.
Body composition analysis
Body composition analysis was conducted as described49,50 with staff members at the metabolism core of the Nutrition and Obesity Research Center at the UCSF. The Lunar PIXImus densitometer (GE Medical Systems) was used to analyse body composition of each mouse using dual energy X-ray absorptiometry technology. In brief, mice were weighed and anaesthetized with avertin before being immobilized on a sticky mat. X-ray measurements were taken and the region of interest was adjusted to ensure the whole mouse was considered for the analysis. Data output provided bone, tissue and fat measurements.
CLAMS metabolism
Metabolic analysis was conducted as previously described49 with staff members at the metabolism core of the Nutrition and Obesity Research Center at the UCSF. In brief, mice were singly housed for one week for habituation to the experimental conditions. Mice were then placed in the CLAMS and monitored for five days. Data were generated in 1 h bins and used to calculate metabolic parameters, including oxygen consumption (\({V}_{{{\rm{O}}}_{2}}\)), carbon dioxide production (\({V}_{{{\rm{CO}}}_{2}}\)), energy expenditure and respiratory exchange ratio.
Morris water maze
Water maze testing was performed as described51,52,53,54. In brief, we filled the water maze pool (diameter, 122 cm) with white opaque water (21° ± 1 °C) and submerged a square 14 cm2 platform 2 cm below the surface. Mice underwent two pretraining trials that consisted of swimming through a channel to mount a rescue platform, before hidden training. The platform was kept in the same submerged spot during all hidden platform training trials; the location where mice were dropped into the pool varied between trials. For the hidden trials, mice received four trials daily for eight days. For the probe trials, the platform was removed, mice were allowed to swim for 60 s, and their latency to enter the previous platform area was recorded for young mice whereas the percentage of time spent in the target quadrant was recorded for old mice. In this study, the latency probe measure showed a dynamic range in young mice; by contrast, old mice showed a ceiling effect of the assay. In old mice, the percentage of time spent in the target quadrant was a sensitive probe measure as non-transgenic controls showed clear age-induced impairment whereas young mice showed a ceiling effect. Notably, probe measures and their sensitivities to memory in young and old mice can also vary between water maze studies. Following probe testing, mice were assessed for their ability to find the platform when it is marked with a visible cue (15 cm pole on the platform).
Open field
Open-field testing was carried out as described18. In brief, mice were acclimatized to the room for 1 h before testing and allowed to explore the open field for 10 min. The open field consisted of a clear plastic chamber (41 × 30 cm) and total activity was detected by measuring beam breaks using an automated Flex-Field/Open Field Photobeam Activity System (San Diego Instruments). Visual cues included a fan, wires and a grid, and were located on three walls. For repeat testing, mice were placed in the same chamber. The index of forgetfulness was calculated by subtracting activity levels on the first day of testing in middle and old age from activity levels on the last day of testing of the previous life stage.
EPM
EPM testing was carried out as described51. The room was maintained in dim light for both habituation and testing. In brief, mice were habituated to the testing room for 1 h before testing. Mice were placed in the centre of the EPM facing the open arm and allowed to explore for 10 min. Distance travelled and the percentage of time spent in the open versus closed arms was recorded using the Kinder Scientific Elevated Plus Maze and MotorMonitor system.
Two-trial large Y maze
The two-trial large Y maze test (with visual cues at the end of each arm) was carried out as described53,55. Sixteen hours after a training session during which the novel arm was closed off, mice were returned to the two-trial large Y maze and allowed to explore all arms freely for 5 min. Time spent in the novel and familiar arm was recorded using the AnyMaze software and the novel to familiar ratio was calculated.
Novel place recognition
Testing was carried out as described56. In brief, mice were acclimatized to the testing room for 1 h before testing, which was performed in a square white chamber (40 × 40 cm) under dim lighting. During the training session, mice were presented with two identical objects placed equidistant from each other and from the surrounding chamber walls. During this training session, mice showed a similar preference for each of the objects. For the test session 4 h later, one of the objects was moved to a new location and mice were allowed to explore for 10 min. Time of object exploration was obtained from the videos using the CleverSys TopScan Automated Behavior Analysis System (v.3.0) and analysed.
Epigenetic DNA age analysis
Hippocampal tissue samples were flash-frozen. Samples then underwent sample library preparation and sequencing analysis as described (Zymo Research)57. In brief, genomic DNA was extracted using the Quick-DNA Miniprep plus kit and bisulfite converted using the EZ DNA Methylation Lightning kit. The samples were then enriched for sequencing of over 500 age-associated gene loci on an Illumina HiSeq 1500 instrument using 100-bp paired-end sequencing (Xm and Xm+Xp hippocampal samples) or Illumina NovaSeq 6000 instrument using 150-bp paired-end sequencing (Xm and Xp hippocampal neurons; Xm and Xm+Xp blood samples). Illumina’s base calling software was used to identify sequence reads and aligned to a reference genome using Bismark, an aligner optimized for bisulfite sequence calling (http://www.bioinformatics.babraham.ac.uk/projects/bismark/). The methylation level was determined by proportion of the numbers of ‘C’ reported to the total numbers of ‘C’ and ‘T’. Calculated DNA methylation values obtained from the sequence data were used to predict the epigenetic age using a proprietary DNAge predictor (Zymo).
FACS
Fresh hippocampal tissue was first homogenized into single-cell suspension as previously described58. We enriched for neurons by applying the cell suspension to an Optiprep density gradient and collected only the neuronal fraction for FACS sorting58. Pre-enrichment for neurons through a density gradient was performed prior to FACS sorting of neurons for the young and old RNA-seq study, and not for the replicate RNA-seq study in young mice or the epigenetic analysis. For parent-of-X origin analysis, hippocampal cells were separated into Xm active and Xp active cells using a Sony SH800 FACS machine with a 100-mm cartridge at 40 psi. Cells were collected into a 15 ml flacon tube containing 2 ml sample collection buffer. Samples and collection tubes for sorting were kept at 4 °C during sorting. Hippocampi from six mice were pooled together to form a single sample. In total, five samples derived from 30 mice were prepared. Each sample was FACS-sorted into Xm cells (GFP+, green) and Xp cells (tdTomato+, red). For CRISPRa+ samples, we sorted dCas9+ and GFP+ nuclei using a 100 mm nozzle at 100 psi on a BD Biosciences FACSAria III. For both experiments, samples collected after sorting were centrifuged at 1,000 rpm for 10 min. The supernatant was discarded, and cells were resuspended in 250 ml of Trizol and stored at −80 °C until RNA-seq sample preparation and analysis.
Neuronal nucleus isolation
Neuronal nuclei were isolated using the Nuclei EZ Prep Isolation kit (Sigma NUC-101). Frozen hippocampi were thawed on ice for 25 min before addition of 2 ml of ice-cold EZ lysis buffer. A hand-held homogenizer was used to completely homogenize the tissue while avoiding frothing. An additional 2 ml of ice-cold EZ lysis buffer was added and samples were then incubated on ice for 10 min. Samples were then centrifuged at 500g for 10 min at 4 °C. The supernatant was discarded and the pellet was resuspended in 1 ml ice-cold EZ lysis buffer. Once the pellet was properly resuspended, an additional 3 ml of ice-cold EZ lysis buffer was added and samples were incubated on ice for 15 min. Samples were then centrifuged at 500g for 10 min at 4 °C. The supernatant was discarded, and the pellet was resuspended in 350 ml of EZ storage buffer. Nuclei were filtered through a 30-mm cell strainer (MACS 130-041-407), counted and stored at −80 °C until FACS sorting.
RNA-seq
In brief, RNA sequencing libraries were prepared using the SMART-Seq v4Ultra Low Input RNA Kit (Clontech). Paired-end reads were obtained using an Illumina HiSeq instrument. The quality of the reads was determined using FastQC, and more than 90% of reads from each sample had a mean quality score over 30. The trimmed reads were mapped to the Mus musculus GRCm38 reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. Unique gene hit counts from exons were calculated using featureCounts in the Subread package v.1.5.2. Downstream differential expression analysis was performed using DESeq2. Sequencing was performed by Azenta Life Sciences.
Identification of imprinted genes and RT–qPCR validation
Imprinted genes were selected on the basis of a list of criteria as follows: (1) significant P value and adjusted P value; (2) mean of normalized gene expression from one sample group of less than 50 and mean of normalized gene expression from the other sample group of over 100; (3) significant χ2 P value and adjusted significant χ2 P value; and (4) fold change above 10. On the basis of these criteria, we identified five imprinted genes. RT–qPCR was used to validate the expression of imprinted genes identified in the RNA-seq analysis. Primers were designed using the NCBI Primer Blast page and purchased from Integrated DNA Technologies. Sash3 Fwd, CTGGCAGTGAAGAGGCTGAA, Rev, GACCCTGCAGTTGCTCTTCT; Cysltr1 Fwd, GGTACCAGATAGAGGTCTCCC, Rev, CTCCAGGAATGTCTGCTTGGT; Tlr13 Fwd, TCCTCCCTCCCTGGAGTTTT, Rev, AGGCACCTTCGTCGATCTTC; Tlr7 Fwd, TGCACTCTTCGCAGCAACTA, Rev, ATGTCTCTTGCTGCCCCAAA; Xlr3b Fwd, AAAAGGAAGGCCACTGACAC, Rev, ACCAGCATCAAGGACTTCTCTG; Gapdh Fwd, GGGAAGCCCATCACCATCTT, Rev, GCCTTCTCCATGGTGGTGAA; 18S RNA Fwd, AGGGGAGAGCGGGTAAGAGA, Rev: GGACAGGACTAGGCGGAACA.
Lentivirus production and stereotaxic injection
Simultaneous overexpression of Sash3, Tlr7 and Cysltr1 was achieved using a dCas9 Synergistic Activation Mediator Lentivirus (Lenti-hSyn-dCas9-VP64-p65-RTA-NLS-SV40-Puro virus) and a lentivirus containing sgRNAs for the three genes (Lenti-U6-Sash3 sgRNA-H1-Cysltr1 sgRNA-U6-Tlr7 sgRNA2-PGK-GFP). Two sets of sgRNAs were pooled together to improve efficiency of gene upregulation (Extended Data Table 2). A similar construct with a scrambled sequence (Lenti-U6-Scrambled sgRNA-PGK-GFP) was used as control (Extended Data Table 2). Active lentiviral particles were obtained from Applied Biological Materials (ABM). ABM performed lentiviral packaging in HEK293T cells using a second-generation co-transfection system. HEK293T cells were subcultured at 70% density in a 15-cm dish one day before virus production. The following day, transfection was performed using 180 μg total plasmid DNA (60 μg expression vector, 120 μg second-generation packaging mix (ABM LV003) and 80 µl lentifectin (ABM G2500)) in the absence of serum for 5 h before restoring the culture conditions back to DMEM + 5% FBS. Then 72 h after transfection, supernatant viruses were collected, purified and stored in PBS storage buffer. The final recombinant lentivirus were validated by titre and HEK293T transduction to ensure the virus were free from bacteria and mycoplasma contamination. Next 18-month-old C57BL6 wild-type mice were anaesthetized using isoflurane at 2–3% and placed in a stereotaxic frame. Then 5 µl of lentiviral vectors (2.5 μl dCas9 + 2.5 µl sgRNA per hemisphere) were stereotactically injected bilaterally into the dentate gyrus of the hippocampus using the coordinates, anteroposterior = −2.1, mediolateral = ±1.7 and dorsoventral = 1.9. After surgery, mice were allowed to wake up completely on a heating pad before being returned to their home cage. All behavioural assays were conducted beginning at 4 weeks and ending at 16 weeks after lentiviral injections.
Immunofluorescence microscopy
Immunofluorescence was performed as previously described59. In brief, mice were perfused with cold PBS (10 ml min−1) for 5 min using a peristaltic pump. Whole brains were then collected and post-fixed in 4% paraformaldehyde for 48 h and subsequently preserved in 30% sucrose (prepared in PBS). Whole brains were sectioned coronally at 40 μm thickness on a freezing sliding microtome throughout the entire hippocampus. Sections were stored in the cryoprotective medium at −20 °C. Free-floating sections were blocked with donkey serum and incubated with primary antibodies at 4 °C overnight at the following concentration for microscopy: rabbit anti-GFP (1:1,000, Sigma G1544) and mouse anti-dCas9 (1:500, Invitrogen MA523519). After washing, sections were incubated with donkey anti-rabbit Alexa Fluor 488 (1:1,000, Thermo Fisher, A32790) and donkey anti-mouse Alexa Fluor 594 (1:1,000, Thermo Fisher, A21203) at room temperature for 2 h. DAPI (300 nM) was added during the last 10 min of the 2 h incubation at room temperature. Sections were washed and mounted with Vectashield before imaging on digital fluorescence microscope with spinning-disk confocal system (Nikon CSU-W1).
Statistical analysis
Experimenters were blinded to genotype. Statistical analyses were carried out using GraphPad Prism (v.7.0) for t-tests and two-way ANOVAs and R Studio (v.2.0) for mixed-model ANOVAs and post-hoc tests. All tests were two tailed unless indicated otherwise. Differences between two means were assessed using unpaired t-tests and a two-way ANOVA to assess differences among multiple means for all experiments unless otherwise stated. Post-hoc tests were conducted with Bonferroni–Holm correction in R to control for a family-wise error rate at α = 0.05 when rounded to two decimal points, unless indicated otherwise. A mixed-model ANOVA was used to analyse Morris water maze and open-field data and included effects for repeated measures. Exclusion criteria (greater than 2 s.d. above or below the mean) were defined a priori to ensure unbiased exclusion of outliers in mouse behaviour studies. Error bars represent the s.e.m. and null hypotheses were rejected at or below a P value of 0.05 when rounded to two decimal points. Linear models were fitted in R using the standard lme package.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.